Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma
Abstract
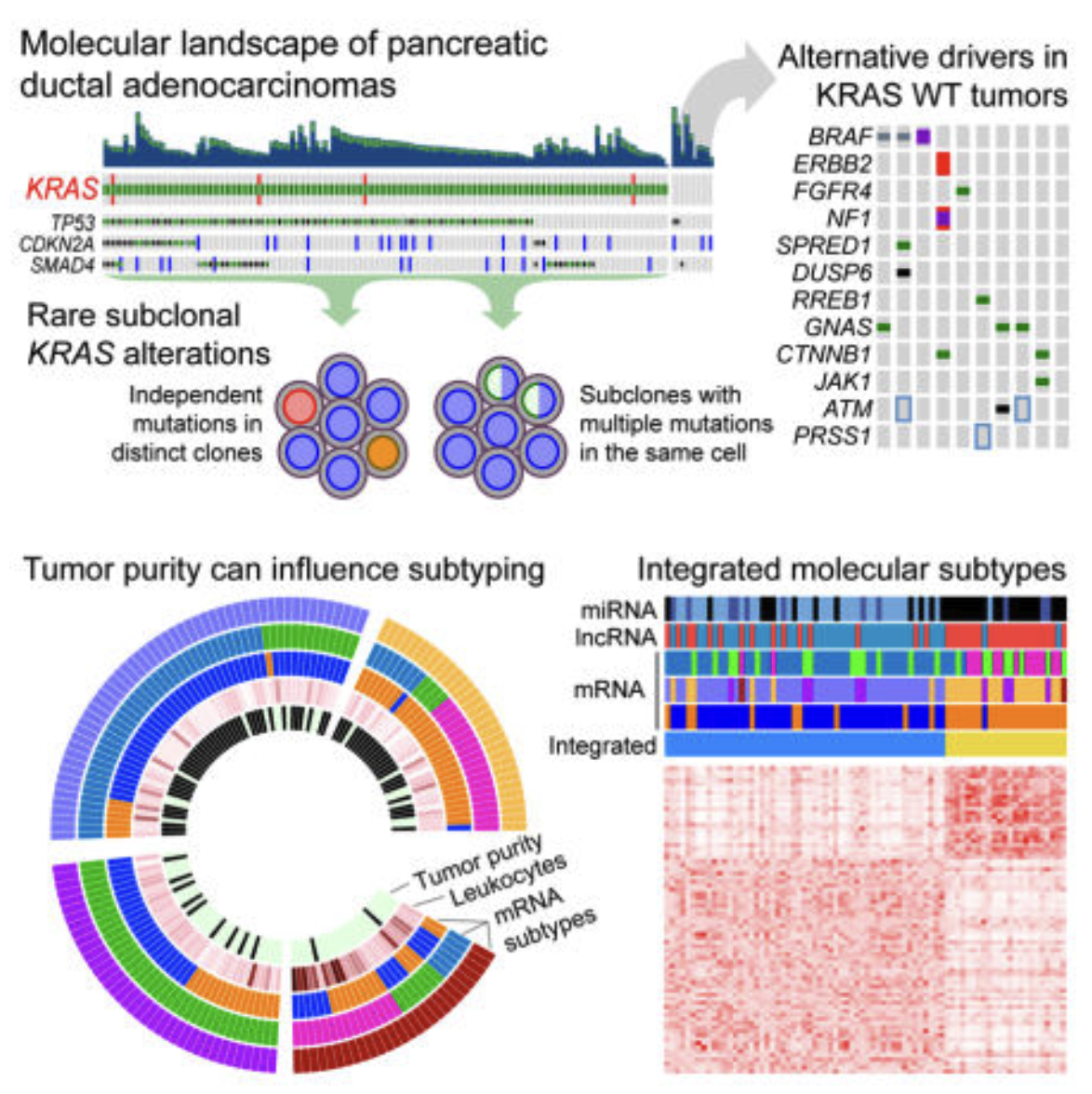
We performed integrated genomic, transcriptomic, and proteomic profiling of 150 pancreatic ductal adenocarcinoma (PDAC) specimens, including samples with characteristic low neoplastic cellularity. Deep whole-exome sequencing revealed recurrent somatic mutations in KRAS, TP53, CDKN2A, SMAD4, RNF43, ARID1A, TGFβR2, GNAS, RREB1, and PBRM1. KRAS wild-type tumors harbored alterations in other oncogenic drivers, including GNAS, BRAF, CTNNB1, and additional RAS pathway genes. A subset of tumors harbored multiple KRAS mutations, with some showing evidence of biallelic mutations. Protein profiling identified a favorable prognosis subset with low epithelial-mesenchymal transition and high MTOR pathway scores. Associations of non-coding RNAs with tumor-specific mRNA subtypes were also identified. Our integrated multi-platform analysis reveals a complex molecular landscape of PDAC and provides a roadmap for precision medicine.
Citation
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma”, Cancer Cell 32(2):185-203.e13 (2017). doi:10.1016/j.ccell.2017.07.007