Guidelines for reporting single-cell RNA-seq experiments
Abstract
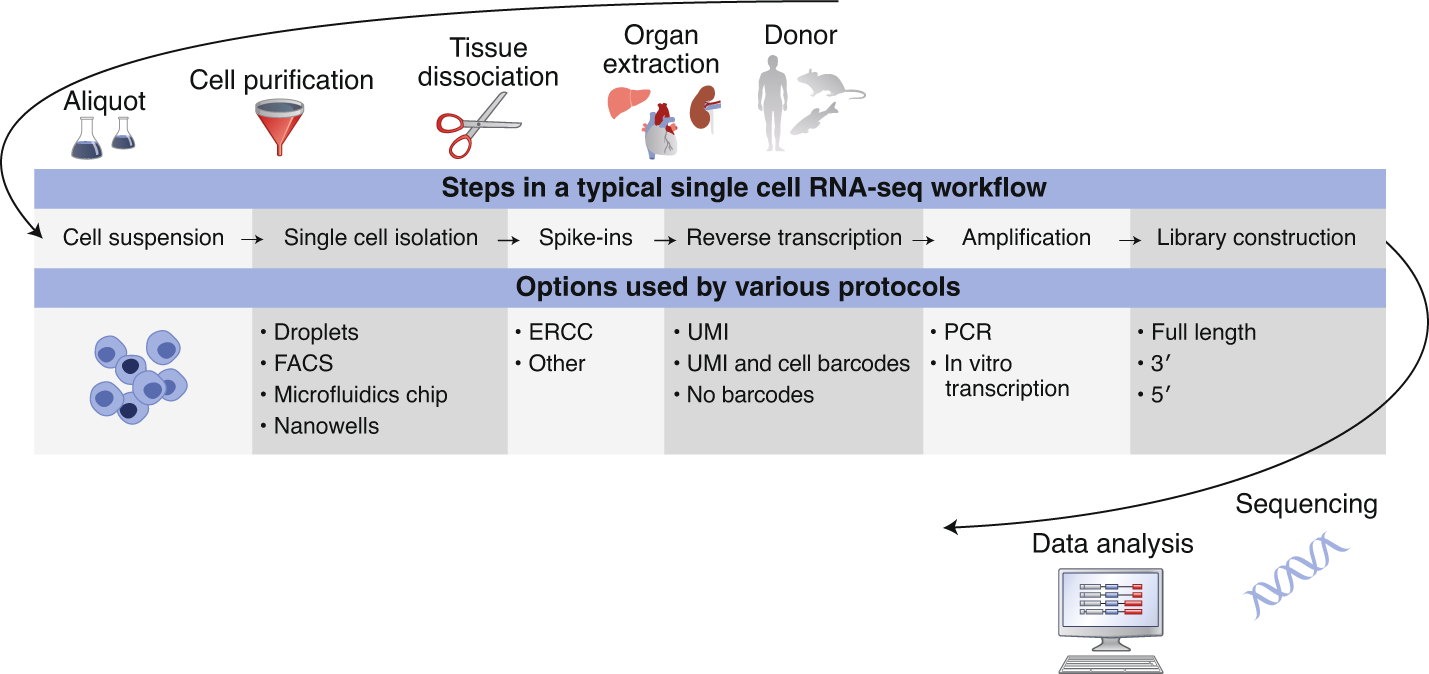
Single-cell RNA-Sequencing (scRNA-Seq) has undergone major technological advances in recent years, enabling the conception of various organism-level cell atlassing projects. With increasing numbers of datasets being deposited in public archives, there is a need to address the challenges of enabling the reproducibility of such data sets. Here, we describe guidelines for a minimum set of metadata to sufficiently describe scRNA-Seq experiments, ensuring reproducibility of data analyses.
Citation
A Füllgrabe, N George, M Green, P Nejad, B Aronow, SK Fexova, C Fischer, MA Freeberg, L Huerta, N Morrison, RH Scheuermann, D Taylor, N Vasilevsky, L Clarke, N Gehlenborg, J Kent, J Marioni, S Teichmann, A Brazma, I Papatheodorou. “Guidelines for reporting single-cell RNA-seq experiments”, Nature Biotechnology 38(12):1384-1386 (2020). doi:10.1038/s41587-020-00744-z