CLIMP-Cluster-based Imputation of Missing Values in Microarray Data
Abstract
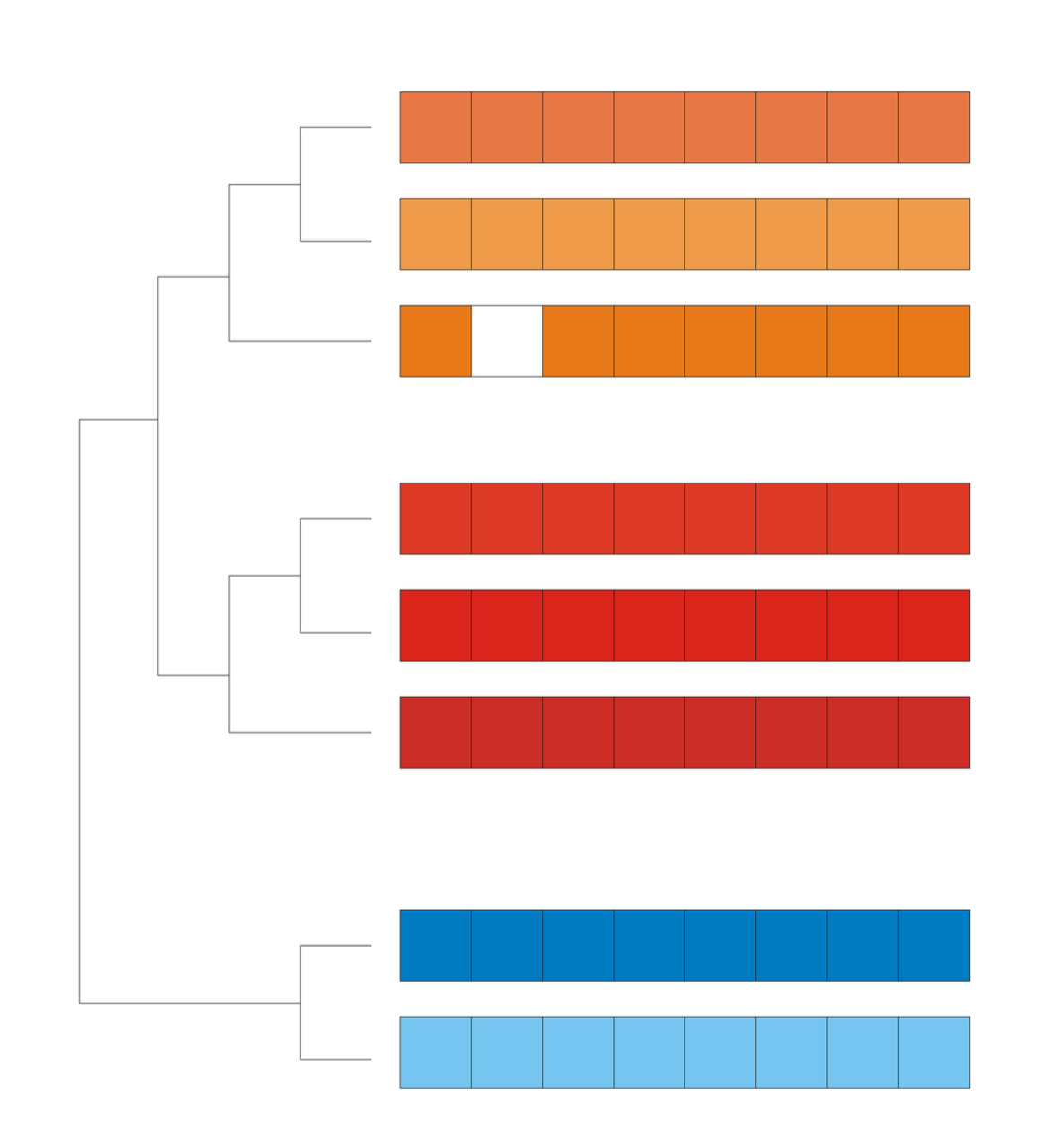
Microarray data often contain missing values due to experimental and technical limitations, which can hinder accurate analysis. CLIMP (Cluster-based Imputation) uses hierarchical clustering to estimate missing values based on gene expression similarity. Compared to existing techniques like KNNimpute and BPCA, CLIMP offers a structured approach to improving data integrity while maintaining comparable accuracy to KNN-based methods.
Citation
N Gehlenborg. “CLIMP-Cluster-based Imputation of Missing Values in Microarray Data”, (2003).