Publications
Preprints | |
|
PJ Van Camp, N Gehlenborg, A Hollander. “HOLLOW TREE: Harnessing Online Learning with Large-language-model Oriented Web-apps for Teaching, Research, Evaluation, and Engagement”, OSF Preprints (2025). doi:10.31219/osf.io/z8gpd_v1 |
|
ML Turner, TC Smits, TS Liaw, B Honick, B Shirey, L Choy, N Akhmetov, S An, D Betancur, D Bordelon, K Burke, I Cao-Berg, J Conroy, C Csonka, P Cuda, S Donahue, S Fisher, D Furst, E Hanna, J Hardi, T Kakar, MS Keller, X Li, Y Ma, A McWilliams, A Money, R Morgan, E Moerth, J Muerto, MA Musen, E Nic, MJ O’Connor, G Phillips, A Ropelewski, R Sablosky, S Saripalli, M Sibilla, D Simmel, A Simmons, X Tang, J Welling, Z Yuan, M Hemberg, M Ruffalo, J Silverstein, P Blood, N Gehlenborg. “HuBMAP Data Portal: A Resource for Multi-Modal Spatial and Single-Cell Data of Healthy Human Tissues”, arXiv (2025). doi:10.48550/ARXIV.2511.05708 |

|
TC Smits, N Akhmetov, TS Liaw, MS Keller, E Mörth, N Gehlenborg. “scellop: A Scalable Redesign of Cell Population Plots for Single-Cell Data”, arXiv (2025). doi:10.48550/ARXIV.2510.09554 |
|
HN Nguyen and N Gehlenborg. “Safire: Similarity Framework for Visualization Retrieval”, 2025 IEEE Visualization and Visual Analytics (VIS). To appear (2025). doi:10.31219/osf.io/p47z5_v4 |

|
HN Nguyen, S L’Yi, TC Smits, S Gao, M Zitnik, N Gehlenborg. “Geranium: Multimodal Retrieval of Genomics Data Visualizations”, OSF Preprints (2025). doi:10.31219/osf.io/zatw9_v5 |
|
E Moerth, B Beinder, R Raidou, N Gehlenborg. “Nexus: Interactive Visual Exploration of 3D Spatial Connectivity for Tissue Analysis”, OSF Preprints (2025). doi:10.31219/osf.io/nypmf_v1 |

|
S Luo, MS Keller, T Kakar, L Choy, N Gehlenborg. “EasyVitessce: auto-magically adding interactivity to Scverse single-cell and spatial biology plots”, arXiv (2025). doi:10.48550/arXiv.2510.19532 |

|
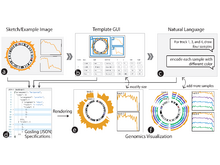
D Lange, S Gao, P Sui, A Money, P Misner, M Zitnik, N Gehlenborg. “YAC: Bridging Natural Language and Interactive Visual Exploration with Generative AI for Biomedical Data Discovery”, arXiv (2025). doi:10.48550/arXiv.2509.19182 |

|
D Lange, P Sui, S Gao, M Zitnik, N Gehlenborg. “DQVis Dataset: Natural Language to Biomedical Visualization”, Advances in Neural Information Processing Systems (NeurIPS). To appear (2025). doi:10.31219/osf.io/rqb7u_v1 |

|
MS Keller, N Lucarelli, Y Chen, S Border, A Janowczyk, J Himmelfarb, M Kretzler, J Hodgin, L Barisoni, D Demeke, L Herlitz, G Moeckel, AZ Rosenberg, Y Ding, Kidney Precision Medicine Project, HuBMAP Consortium, P Sarder, N Gehlenborg. “Interactive visualization of kidney micro-compartmental segmentations and associated pathomics on whole slide images”, arXiv (2025). doi:10.48550/arXiv.2510.19499 |
|
L Weru, S L’Yi, TC Smits, N Gehlenborg. “Using OpenKeyNav to Enhance the Keyboard-Accessibility of Web-based Data Visualization Tools”, OSF Preprints (2024). doi:10.31219/osf.io/3wjsa |
|
Q Wang, X Liu, N Gehlenborg. “What are People Talking about in High-Dimension Data Visualization? LLM-supported Analysis of Domain Literature”, OSF Preprints (2024). doi:10.31219/osf.io/qtsak |

|
V Stoiber, N Gehlenborg, W Aigner, M Streit. “time-i-gram: A Grammar for Interactive Visualization of Time-based Data”, OSF Preprints (2024). doi:10.31219/osf.io/m9ubg |

|
E Mörth, C Nielsen, M Turner, L Xianhao, MS Keller, J Beyer, H Pfister, C Zhu-Tian, N Gehlenborg. “A Mixed Reality and 2D Display Hybrid Approach for Visual Analysis of 3D Tissue Maps”, OSF Preprints (2024). doi:10.31219/osf.io/zka2j |

|
HG Zhang, JP Honerlaw, M Maripuri, MJ Samayamuthu, BR Beaulieu-Jones, HS Baig, S L’Yi, YL Ho, M Morris, VA Panickan, X Wang, C Hong, GM Weber, KP Liao, S Visweswaran, BWQ Tan, W Yuan, N Gehlenborg, S Muralidhar, RB Ramoni, The Consortium for Clinical Characterization of COVID-19 by EHR (4CE), IS Kohane, Z Xia, K Cho, T Cai, GA Brat. “Characterizing the use of the ICD-10 Code for Long COVID in 3 US Healthcare Systems”, medRxiv (2023). doi:10.1101/2023.02.12.23285701 |
|
Q Wang, S L’Yi, N Gehlenborg. “Improving the Utility and Usability of Visualization in AI-driven Scientific Discovery”, OSF Preprints (2022). doi:10.31219/osf.io/hpd6u |
|

|
J Fan, D Fan, K Slowikowski, N Gehlenborg, P Kharchenko. “UBiT2: a client-side web-application for gene expression data analysis”, bioRxiv (2017). doi:10.1101/118992 |
2026 | |
|
D Kouřil, T Manz, T Clarence, N Gehlenborg. “Uchimata: a toolkit for visualization of 3D genome structures on the web and in computational notebooks”, Bioinformatics btag035 (2026). doi:10.1093/bioinformatics/btag035 |

|
D Kouřil, T Manz, S L’Yi, N Gehlenborg. “Design Space and Declarative Grammar for 3D Genomic Data Visualization”, IEEE Transactions on Visualization and Computer Graphics 32(1):1-11 (2026). doi:10.1109/TVCG.2025.3634654 |
2025 | |
|
SS Walters, A Valderrama, TC Smits, D Kouřil, HN Nguyen, S L’Yi, D Lange, N Gehlenborg. “GQVis: A Dataset of Genomics Data Questions and Visualizations for Generative AI”, 2025 IEEE VIS Workshop on GenAI, Agents, and the Future of VIS (VISxGenAI) (2025). doi:10.48550/arXiv.2510.13816 |

|
A Van Den Brandt, S L’Yi, HN Nguyen, A Vilanova, N Gehlenborg. “Understanding Visualization Authoring Techniques for Genomics Data in the Context of Personas and Tasks”, IEEE Transactions on Visualization and Computer Graphics 31(1):1180-1190 (2025). doi:10.1109/TVCG.2024.3456298 |

|
TC Smits, L Weru, N Gehlenborg, S L’Yi. “Ten Simple Rules for Making Biomedical Data Resources Accessible”, PLOS Computational Biology 21(11):e1013657 (2025). doi:10.1371/journal.pcbi.1013657 |
|
D Nakikj, D Kreda, K Luthria, N Gehlenborg. “Patient-Generated Collections for Organizing Electronic Health Record Data to Elevate Personal Meaning, Improve Actionability, and Support Patient–Health Care Provider Communication: Think-Aloud Evaluation Study”, JMIR Human Factors 12:e50331 (2025). doi:10.2196/50331 |

|
E Mörth, Z Kostic, N Gehlenborg, H Pfister, J Beyer, C Nobre. “Beyond Time and Accuracy: Strategies in Visual Problem-Solving”, Proceedings of the 2025 CHI Conference on Human Factors in Computing Systems (2025). doi:10.1145/3706598.3714024 |

|
E Mörth, K Sidak, Z Maliga, T Möller, N Gehlenborg, P Sorger, H Pfister, J Beyer, R Krüger. “Cell2Cell: Explorative Cell Interaction Analysis in Multi-Volumetric Tissue Data”, IEEE Transactions on Visualization and Computer Graphics 31(1):569-579 (2025). doi:10.1109/TVCG.2024.3456406 |

|
T Minten, S Bick, S Adelson, N Gehlenborg, LM Amendola, F Boemer, AJ Coffey, N Encina, A Ferlini, J Kirschner, BE Russell, L Servais, KL Sund, RJ Taft, P Tsipouras, H Zouk, ICoNS Gene List Contributors, D Bick, International Consortium on Newborn Sequencing (ICoNS), RC Green, NB Gold. “Data-driven consideration of genetic disorders for global genomic newborn screening programs”, Genetics in Medicine 27(7):101443 (2025). doi:10.1016/j.gim.2025.101443 |

|
T Manz, F Lekschas, E Greene, G Finak, N Gehlenborg. “A General Framework for Comparing Embedding Visualizations Across Class-Label Hierarchies”, IEEE Transactions on Visualization and Computer Graphics 31(1):283-293 (2025). doi:10.1109/TVCG.2024.3456370 |

|
S L’Yi, A van den Brandt, E Adams, HN Nguyen, N Gehlenborg. “Learnable and Expressive Visualization Authoring through Blended Interfaces”, IEEE Transactions on Visualization and Computer Graphics 31(1):459-469 (2025). doi:10.1109/TVCG.2024.3456598
|

|
S L’Yi, HG Zhang, AP Mar, TC Smits, L Weru, S Rojas, A Lex, N Gehlenborg. “A comprehensive evaluation of life sciences data resources reveals significant accessibility barriers”, Scientific Reports (2025). doi:10.1038/s41598-025-08731-7 |

|
D Lange, S Gao, P Sui, A Money, P Misner, M Zitnik, N Gehlenborg. “A Generative AI System for Biomedical Data Discovery with Grammar-Based Visualizations”, 2025 IEEE VIS Workshop on GenAI, Agents, and the Future of VIS (VISxGenAI) (2025). doi:10.48550/ARXIV.2509.16454 |

|
J Kubica, S Korsak, KH Banecki, D Schirman, AD Yadavalli, A Brenner Clerkin, D Kouřil, M Kadlof, B Busby, D Plewczynski. “The challenge of chromatin model comparison and validation: A project from the first international 4D Nucleome Hackathon”, PLOS Computational Biology 21(8):e1013358 (2025). doi:10.1371/journal.pcbi.1013358 |
|
MS Keller, E Mörth, TC Smits, S Warchol, G Guo, Q Wang, R Krueger, H Pfister, N Gehlenborg. “The State of Single-Cell Atlas Data Visualization in the Biological Literature”, IEEE Computer Graphics and Applications 1-15 (2025). doi:10.1109/MCG.2025.3583979 |

|
J Dekker, BA Oksuz, Y Zhang, Y Wang, MK Minsk, S Kuang, L Yang, JH Gibcus, N Krietenstein, OJ Rando, J Xu, DH Janssens, S Henikoff, A Kukalev, W Andréa, W Winick-Ng, R Kempfer, A Pombo, M Yu, P Kumar, L Zhang, AS Belmont, T Sasaki, T Van Schaik, L Brueckner, D Peric-Hupkes, B Van Steensel, P Wang, H Chai, M Kim, Y Ruan, R Zhang, SA Quinodoz, P Bhat, M Guttman, W Zhao, S Chien, Y Liu, SV Venev, D Plewczynski, II Azcarate, D Szabó, CJ Thieme, T Szczepińska, M Chiliński, K Sengupta, M Conte, A Esposito, A Abraham, R Zhang, Y Wang, X Wen, Q Wu, Y Yang, J Liu, L Boninsegna, A Yildirim, Y Zhan, AM Chiariello, S Bianco, L Lee, M Hu, Y Li, RJ Barnett, AL Cook, DJ Emerson, C Marchal, P Zhao, PJ Park, BH Alver, AJ Schroeder, R Navelkar, C Bakker, W Ronchetti, S Ehmsen, AD Veit, N Gehlenborg, T Wang, D Li, X Wang, M Nicodemi, B Ren, S Zhong, JE Phillips-Cremins, DM Gilbert, KS Pollard, F Alber, J Ma, WS Noble, F Yue. “An integrated view of the structure and function of the human 4D nucleome”, Nature (2025). doi:10.1038/s41586-025-09890-3 |
2024 | |

|
TC Smits, S L’Yi, HN Nguyen, AP Mar, N Gehlenborg. “Explaining Unfamiliar Genomics Data Visualizations to a Blind Individual through Transitions”, 2024 1st Workshop on Accessible Data Visualization (AccessViz) (2024). doi:10.1109/AccessViz64636.2024.00010 |
|
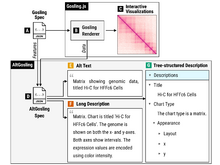
TC Smits, S L’Yi, AP Mar, N Gehlenborg. “AltGosling: Automatic Generation of Text Descriptions for Accessible Genomics Data Visualization”, Bioinformatics btae670 (2024). doi:10.1093/bioinformatics/btae670 |
|
I Sarfraz, Y Wang, A Shastry, WK Teh, A Sokolov, BR Herb, HH Creasy, I Virshup, R Dries, K Degatano, A Mahurkar, DJ Schnell, P Madrigal, J Hilton, N Gehlenborg, T Tickle, JD Campbell. “MAMS: matrix and analysis metadata standards to facilitate harmonization and reproducibility of single-cell data”, Genome Biology 25(1):205 (2024). doi:10.1186/s13059-024-03349-w |
|
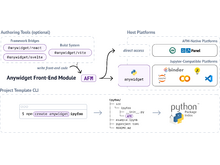
T Manz, N Abdennur, N Gehlenborg. “anywidget: reusable widgets for interactive analysis and visualization in computational notebooks”, Journal of Open Source Software 9(102):6939 (2024). doi:10.21105/joss.06939 |
|
T Manz, N Gehlenborg, N Abdennur. “Any notebook served: authoring and sharing reusable interactive widgets”, Proceedings of the 23rd Python in Science Conference (2024). doi:10.25080/NRPV2311 |

|
L Taing, A Dandawate, S L’Yi, N Gehlenborg, M Brown, CA Meyer. “Cistrome Data Browser: integrated search, analysis and visualization of chromatin data”, Nucleic Acids Research 52(D1):D61-D66 (2024). doi:10.1093/nar/gkad1069 |

|
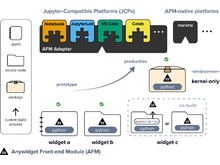
MS Keller, T Manz, N Gehlenborg. “Use-Coordination: Model, Grammar, and Library for Implementation of Coordinated Multiple Views”, 2024 IEEE Visualization and Visual Analytics (VIS) (2024). doi:10.1109/VIS55277.2024.00041 |
|
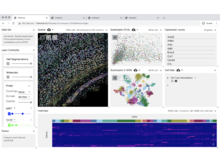
MS Keller, I Gold, C McCallum, T Manz, PV Kharchenko, N Gehlenborg. “Vitessce: a framework for integrative visualization of multi-modal and spatially-resolved single-cell data”, Nature Methods 1-5 (2024). doi:10.1038/s41592-024-02436-x |
|
K Huang, P Chandak, Q Wang, S Havaldar, A Vaid, J Leskovec, GN Nadkarni, BS Glicksberg, N Gehlenborg, M Zitnik. “A foundation model for clinician-centered drug repurposing”, Nature Medicine (2024). doi:10.1038/s41591-024-03233-x |

|
DL Gisch, M Brennan, BB Lake, J Basta, MS Keller, R Melo Ferreira, S Akilesh, R Ghag, C Lu, YH Cheng, KS Collins, SV Parikh, BH Rovin, L Robbins, L Stout, KY Conklin, D Diep, B Zhang, A Knoten, D Barwinska, M Asghari, AR Sabo, MJ Ferkowicz, TA Sutton, KJ Kelly, IH De Boer, SE Rosas, K Kiryluk, JB Hodgin, F Alakwaa, S Winfree, N Jefferson, A Türkmen, JP Gaut, N Gehlenborg, CL Phillips, TM El-Achkar, PC Dagher, T Hato, K Zhang, J Himmelfarb, M Kretzler, S Mollah, the Kidney Precision Medicine Project (KPMP), B Lake, A Morales, I Stillman, S Lecker, S Bogen, A Verma, G Yu, I Schmidt, J Henderson, L Beck, P Yadati, S Waikar, AA Amodu, S Maikhor, T Ilori, MR Colona, A Weins, G McMahon, N Hacohen, A Greka, JL Marshall, PJ Hoover, VS Viswanathan, D Crawford, M Aulisio, W Bush, Y Chen, A Madabhushi, C O’Malley, C Gadegbeku, D Sendrey, E Poggio, J O’Toole, J Sedor, J Taliercio, L Bush, L Herlitz, E Palmer, J Nguyen, K Spates-Harden, L Cooperman, S Jolly, C Vinovskis, A Bomback, J Barasch, K Kiryluk, P Appelbaum, V D’Agati, C Berrouet, K Mehl, M Sabatello, N Shang, O Balderes, PA Canetta, S Kudose, J De Pinho Gonçalves, L Migas, R Van De Plas, R Lardenoije, L Barisoni, H Rennke, A Verdoes, A Sabo, DL Gisch, J Williams, K Kelly, K Dunn, M Eadon, M Ferkowicz, P Dagher, S Winfree, S Bledsoe, S Wofford, T Sutton, W Bowen, A Slade, E Record, Y Cheng, Y Jain, B Herr, E Quardokus, A Wang, CPC Villalobos, C Parikh, M Atta, S Menez, Y Wen, A Xu, L Bernard, C Johansen, S Chen, S Rosas, I Donohoe, J Sun, R Knight, A Shpigel, J Bebiak, J Saul, J Ardayfio, R Koewler, R Pinkeney, T Campbell, E Azeloglu, G Nadkarni, J He, J Tokita, K Campbell, M Patel, S Lefferts, SR Iyengar, S Ward, S Coca, C He, Y Xiong, P Prasad, B Rovin, JP Shapiro, S Parikh, SM Madhavan, J Lukowski, D Velickovic, L Pasa-Tolic, G Oliver, O Troyanskaya, R Sealfon, W Mao, A Wong, A Pollack, Y Goltsev, B Ginley, B Lutnick, G Nolan, K Anjani, T Mukatash, ZG Laszik, B Campos, B Thajudeen, D Beyda, E Bracamonte, F Brosius, G Woodhead, K Mendoza, N Marquez, R Scott, R Tsosie, M Saunders, A Rike, ES Woodle, PJ Lee, RR Alloway, T Shi, E Hsieh, J Kendrick, J Thurman, J Wrobel, L Pyle, P Bjornstad, N Lucarelli, P Sarder, A Renteria, A Ricardo, A Srivastava, D Redmond, E Carmona-Powell, J Bui, J Lash, M Fox, N Meza, R Gaba, S Setty, T Kelly, C Lienczewski, D Demeke, E Otto, H Ascani, J Hodgin, J Schaub, J Hartman, L Mariani, M Bitzer, M Rose, N Bonevich, N Conser, P McCown, R Dull, R Menon, R Reamy, S Eddy, U Balis, V Blanc, V Nair, YO He, Z Wright, B Steck, J Luo, R Frey, A Coleman, D Henderson-Brown, J Berge, ML Caramori, O Adeyi, P Nachman, S Safadi, S Flanagan, S Ma, S Klett, S Wolf, T Harindhanavudhi, V Rao, A Mottl, A Froment, E Zeitler, P Bream, S Kelley, M Rosengart, M Elder, P Palevsky, R Murugan, DE Hall, F Bender, J Winters, JA Kellum, M Gilliam, M Tublin, R Tan, G Zhang, K Sharma, M Venkatachalam, A Hendricks, A Kermani, J Torrealba, M Vazquez, N Wang, Q Cai, RT Miller, S Ma, S Hedayati, A Hoofnagle, A Wangperawong, A Berglund, AL Dighe, B Young, B Larson, B Berry, C Alpers, C Limonte, C Stutzke, G Roberts, I De Boer, J Snyder, J Phuong, J Carson, K Rezaei, K Tuttle, K Brown, K Blank, N Sarkisova, N Jefferson, R McClelland, S Mooney, Y Nam, A Wilcox, C Park, F Dowd, K Williams, SM Grewenow, S Daniel, S Shankland, A Pamreddy, H Ye, R Montellano, S Bansal, A Pillai, D Zhang, H Park, J Patel, K Sambandam, M Basit, N Wen, OW Moe, RD Toto, SC Lee, K Sharman, RM Caprioli, A Fogo, J Allen, J Spraggins, K Djambazova, M De Caestecker, M Dufresne, M Farrow, A Vijayan, B Minor, G Nwanne, J Gaut, K Conlon, M Kaushal, SM Diettman, AM Victoria Castro, D Moledina, FP Wilson, G Moeckel, L Cantley, M Shaw, V Kakade, T Arora, S Jain, M Rauchman, MT Eadon. “The chromatin landscape of healthy and injured cell types in the human kidney”, Nature Communications 15(1):433 (2024). doi:10.1038/s41467-023-44467-6 |
2023 | |

|
Q Wang, S L’Yi, N Gehlenborg. “Drava: Concept-Driven Exploration of Small Multiples using Interpretable Latent Vectors”, Proceedings of the 2023 CHI Conference on Human Factors in Computing Systems (2023). doi:10.1145/3544548.3581127 |
|
Q Wang, X Liu, MQ Liang, S L’Yi, N Gehlenborg. “Enabling Multimodal User Interactions for Genomics Visualization Creation”, 2023 IEEE Visualization and Visual Analytics (VIS) (2023). doi:10.1109/VIS54172.2023.00031 |

|
BWL Tan, BWQ Tan, ALM Tan, ER Schriver, A Gutiérrez-Sacristán, P Das, W Yuan, MR Hutch, N García Barrio, M Pedrera Jimenez, N Abu-el-rub, M Morris, B Moal, G Verdy, K Cho, YL Ho, LP Patel, A Dagliati, A Neuraz, JG Klann, AM South, S Visweswaran, DA Hanauer, SE Maidlow, M Liu, DL Mowery, A Batugo, A Makoudjou, P Tippmann, D Zöller, GA Brat, Y Luo, P Avillach, R Bellazzi, L Chiovato, A Malovini, V Tibollo, MJ Samayamuthu, P Serrano Balazote, Z Xia, NHW Loh, L Chiudinelli, CL Bonzel, C Hong, HG Zhang, GM Weber, IS Kohane, T Cai, GS Omenn, JH Holmes, KY Ngiam, JR Aaron, G Agapito, A Albayrak, G Albi, M Alessiani, A Alloni, DF Amendola, F Angoulvant, LLLJ Anthony, BJ Aronow, F Ashraf, A Atz, P Avillach, VA Panickan, PS Azevedo, J Balshi, A Batugo, BK Beaulieu-Jones, BR Beaulieu-Jones, DS Bell, A Bellasi, R Bellazzi, V Benoit, M Beraghi, JL Bernal-Sobrino, M Bernaux, R Bey, S Bhatnagar, A Blanco-Martínez, M Boeker, CL Bonzel, J Booth, S Bosari, FT Bourgeois, RL Bradford, GA Brat, S Bréant, NW Brown, R Bruno, WA Bryant, M Bucalo, E Bucholz, A Burgun, T Cai, M Cannataro, A Carmona, AM Cattelan, C Caucheteux, J Champ, J Chen, KY Chen, L Chiovato, L Chiudinelli, K Cho, JJ Cimino, TK Colicchio, S Cormont, S Cossin, JB Craig, JL Cruz-Bermúdez, J Cruz-Rojo, A Dagliati, M Daniar, C Daniel, P Das, B Devkota, A Dionne, R Duan, J Dubiel, SL DuVall, L Esteve, H Estiri, S Fan, RW Follett, T Ganslandt, N García-Barrio, LX Garmire, N Gehlenborg, EJ Getzen, A Geva, TG González, T Gradinger, A Gramfort, R Griffier, N Griffon, O Grisel, A Gutiérrez-Sacristán, PH Guzzi, L Han, DA Hanauer, C Haverkamp, DY Hazard, B He, DW Henderson, M Hilka, YL Ho, JH Holmes, JP Honerlaw, C Hong, KM Huling, MR Hutch, RW Issitt, AS Jannot, V Jouhet, R Kavuluru, MS Keller, CJ Kennedy, KF Kernan, DA Key, K Kirchoff, JG Klann, IS Kohane, ID Krantz, D Kraska, AK Krishnamurthy, S L’Yi, TT Le, J Leblanc, G Lemaitre, L Lenert, D Leprovost, M Liu, NH Will Loh, Q Long, S Lozano-Zahonero, Y Luo, KE Lynch, S Mahmood, SE Maidlow, A Makoudjou, S Makwana, A Malovini, KD Mandl, C Mao, A Maram, M Maripuri, P Martel, MR Martins, JS Marwaha, AJ Masino, M Mazzitelli, DR Mazzotti, A Mensch, M Milano, MF Minicucci, B Moal, TM Ahooyi, JH Moore, C Moraleda, JS Morris, M Morris, KL Moshal, S Mousavi, DL Mowery, DA Murad, SN Murphy, TP Naughton, CT Breda Neto, A Neuraz, J Newburger, KY Ngiam, WFM Njoroge, JB Norman, J Obeid, MP Okoshi, KL Olson, GS Omenn, N Orlova, BD Ostasiewski, NP Palmer, N Paris, LP Patel, M Pedrera-Jiménez, AC Pfaff, ER Pfaff, D Pillion, S Pizzimenti, T Priya, HU Prokosch, RA Prudente, A Prunotto, V Quirós-González, RB Ramoni, M Raskin, S Rieg, G Roig-Domínguez, P Rojo, P Rubio-Mayo, P Sacchi, C Sáez, E Salamanca, MJ Samayamuthu, LN Sanchez-Pinto, A Sandrin, N Santhanam, JCC Santos, FJ Sanz Vidorreta, M Savino, ER Schriver, P Schubert, J Schuettler, L Scudeller, NJ Sebire, P Serrano-Balazote, P Serre, A Serret-Larmande, M Shah, ZS Hossein Abad, D Silvio, P Sliz, J Son, C Sonday, AM South, F Sperotto, A Spiridou, ZH Strasser, ALM Tan, BWQ Tan, BWL Tan, SE Tanni, DM Taylor, AI Terriza-Torres, V Tibollo, P Tippmann, EMS Toh, C Torti, EM Trecarichi, AK Vallejos, G Varoquaux, ME Vella, G Verdy, JJ Vie, S Visweswaran, M Vitacca, KB Wagholikar, LR Waitman, X Wang, D Wassermann, GM Weber, M Wolkewitz, S Wong, Z Xia, X Xiong, Y Ye, N Yehya, W Yuan, JM Zachariasse, JJ Zahner, A Zambelli, HG Zhang, D Zöller, V Zuccaro, C Zucco. “Long-term kidney function recovery and mortality after COVID-19-associated acute kidney injury: an international multi-centre observational cohort study”, eClinicalMedicine 55:101724 (2023). doi:10.1016/j.eclinm.2022.101724 |

|
F Sperotto, A Gutiérrez-Sacristán, S Makwana, X Li, VN Rofeberg, T Cai, FT Bourgeois, GS Omenn, DA Hanauer, C Sáez, CL Bonzel, E Bucholz, A Dionne, MD Elias, N García-Barrio, TG González, RW Issitt, KF Kernan, J Laird-Gion, SE Maidlow, KD Mandl, TM Ahooyi, C Moraleda, M Morris, KL Moshal, M Pedrera-Jiménez, MA Shah, AM South, A Spiridou, DM Taylor, G Verdy, S Visweswaran, X Wang, Z Xia, JM Zachariasse, JR Aaron, A Adam, G Agapito, A Albayrak, G Albi, M Alessiani, A Alloni, DF Amendola, F Angoulvant, LL Anthony, BJ Aronow, F Ashraf, A Atz, P Avillach, VA Panickan, PS Azevedo, R Badenes, J Balshi, A Batugo, BR Beaulieu-Jones, BK Beaulieu-Jones, DS Bell, A Bellasi, R Bellazzi, V Benoit, M Beraghi, JL Bernal-Sobrino, M Bernaux, R Bey, S Bhatnagar, A Blanco-Martínez, M Boeker, CL Bonzel, J Booth, S Bosari, FT Bourgeois, RL Bradford, GA Brat, S Bréant, NW Brown, R Bruno, WA Bryant, M Bucalo, E Bucholz, A Burgun, T Cai, M Cannataro, A Carmona, AM Cattelan, C Caucheteux, J Champ, J Chen, KY Chen, L Chiovato, L Chiudinelli, K Cho, JJ Cimino, TK Colicchio, S Cormont, S Cossin, JB Craig, JL Cruz-Bermúdez, J Cruz-Rojo, A Dagliati, M Daniar, C Daniel, P Das, B Devkota, A Dionne, R Duan, J Dubiel, SL DuVall, L Esteve, H Estiri, S Fan, RW Follett, T Ganslandt, N García-Barrio, LX Garmire, N Gehlenborg, EJ Getzen, A Geva, RS Goh, TG González, T Gradinger, A Gramfort, R Griffier, N Griffon, O Grisel, A Gutiérrez-Sacristán, PH Guzzi, L Han, DA Hanauer, C Haverkamp, DY Hazard, B He, DW Henderson, M Hilka, YL Ho, JH Holmes, JP Honerlaw, C Hong, KM Huling, MR Hutch, RW Issitt, AS Jannot, V Jouhet, MK Kainth, KF Kate, R Kavuluru, MS Keller, CJ Kennedy, KF Kernan, DA Key, K Kirchoff, JG Klann, IS Kohane, ID Krantz, D Kraska, AK Krishnamurthy, S L’Yi, J Leblanc, G Lemaitre, L Lenert, D Leprovost, M Liu, NH Will Loh, Q Long, S Lozano-Zahonero, Y Luo, KE Lynch, S Mahmood, SE Maidlow, A Makoudjou, S Makwana, A Malovini, KD Mandl, C Mao, A Maram, M Maripuri, P Martel, MR Martins, JS Marwaha, AJ Masino, M Mazzitelli, DR Mazzotti, A Mensch, M Milano, MF Minicucci, B Moal, TM Ahooyi, JH Moore, C Moraleda, JS Morris, M Morris, KL Moshal, S Mousavi, DL Mowery, DA Murad, SN Murphy, TP Naughton, CT Breda Neto, A Neuraz, J Newburger, KY Ngiam, WF Njoroge, JB Norman, J Obeid, MP Okoshi, KL Olson, GS Omenn, N Orlova, BD Ostasiewski, NP Palmer, N Paris, LP Patel, M Pedrera-Jiménez, AC Pfaff, ER Pfaff, D Pillion, S Pizzimenti, T Priya, HU Prokosch, RA Prudente, A Prunotto, V Quirós-González, RB Ramoni, M Raskin, S Rieg, G Roig-Domínguez, P Rojo, N Romero-Garcia, P Rubio-Mayo, P Sacchi, C Sáez, E Salamanca, MJ Samayamuthu, LN Sanchez-Pinto, A Sandrin, N Santhanam, JCC Santos, FJ Sanz Vidorreta, M Savino, ER Schriver, P Schubert, J Schuettler, L Scudeller, NJ Sebire, P Serrano-Balazote, P Serre, A Serret-Larmande, MA Shah, ZS Hossein Abad, D Silvio, P Sliz, J Son, C Sonday, AM South, F Sperotto, A Spiridou, ZH Strasser, AL Tan, BWQ Tan, BWL Tan, SE Tanni, DM Taylor, AI Terriza-Torres, V Tibollo, P Tippmann, EM Toh, C Torti, EM Trecarichi, AK Vallejos, G Varoquaux, ME Vella, G Verdy, JJ Vie, S Visweswaran, M Vitacca, KB Wagholikar, LR Waitman, X Wang, D Wassermann, GM Weber, M Wolkewitz, S Wong, Z Xia, X Xiong, Y Ye, N Yehya, W Yuan, JM Zachariasse, JJ Zahner, A Zambelli, HG Zhang, D Zöller, V Zuccaro, C Zucco, JW Newburger, P Avillach. “Clinical phenotypes and outcomes in children with multisystem inflammatory syndrome across SARS-CoV-2 variant eras: a multinational study from the 4CE consortium”, eClinicalMedicine 64:102212 (2023). doi:10.1016/j.eclinm.2023.102212 |
|
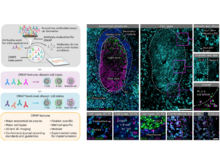
EM Quardokus, DC Saunders, E McDonough, JW Hickey, C Werlein, C Surrette, P Rajbhandari, AM Casals, H Tian, L Lowery, EK Neumann, F Björklund, TV Neelakantan, J Croteau, AE Wiblin, J Fisher, AJ Livengood, KG Dowell, JC Silverstein, JM Spraggins, GS Pryhuber, G Deutsch, F Ginty, GP Nolan, S Melov, D Jonigk, MA Caldwell, IS Vlachos, W Muller, N Gehlenborg, BR Stockwell, E Lundberg, MP Snyder, RN Germain, JM Camarillo, NL Kelleher, K Börner, AJ Radtke. “Organ Mapping Antibody Panels: a community resource for standardized multiplexed tissue imaging”, Nature Methods 20(8):1174-1178 (2023). doi:10.1038/s41592-023-01846-7 |
|
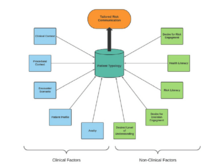
J Panton, BR Beaulieu-Jones, JS Marwaha, AP Woods, D Nakikj, N Gehlenborg, GA Brat. “How surgeons use risk calculators and non-clinical factors for informed consent and shared decision making: A qualitative study”, The American Journal of Surgery 226(5):660-667 (2023). doi:10.1016/j.amjsurg.2023.07.017 |
|
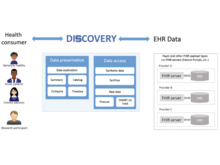
D Nakikj, D Kreda, N Gehlenborg. “New Ways for Patients to Make Sense of Their Electronic Health Record Data Using the Discovery Web Application: Think-Aloud Evaluation Study”, JMIR Formative Research 7:e41346 (2023). doi:10.2196/41346 |
|
D Nakikj, D Kreda, N Gehlenborg. “Alerts and Collections for Automating Patients’ Sensemaking and Organizing of Their Electronic Health Record Data for Reflection, Planning, and Clinical Visits: Qualitative Research-Through-Design Study”, JMIR Human Factors 10:e41552 (2023). doi:10.2196/41552 |
|
J Moore, D Basurto-Lozada, S Besson, J Bogovic, J Bragantini, EM Brown, JM Burel, X Casas Moreno, G De Medeiros, EE Diel, D Gault, SS Ghosh, I Gold, YO Halchenko, M Hartley, D Horsfall, MS Keller, M Kittisopikul, G Kovacs, A Küpcü Yoldaş, K Kyoda, A Le Tournoulx De La Villegeorges, T Li, P Liberali, D Lindner, M Linkert, J Lüthi, J Maitin-Shepard, T Manz, L Marconato, M McCormick, M Lange, K Mohamed, W Moore, N Norlin, W Ouyang, B Özdemir, G Palla, C Pape, L Pelkmans, T Pietzsch, S Preibisch, M Prete, N Rzepka, S Samee, N Schaub, H Sidky, AC Solak, DR Stirling, J Striebel, C Tischer, D Toloudis, I Virshup, P Walczysko, AM Watson, E Weisbart, F Wong, KA Yamauchi, O Bayraktar, BA Cimini, N Gehlenborg, M Haniffa, N Hotaling, S Onami, LA Royer, S Saalfeld, O Stegle, FJ Theis, JR Swedlow. “OME-Zarr: a cloud-optimized bioimaging file format with international community support”, Histochemistry and Cell Biology 160(3):223-251 (2023). doi:10.1007/s00418-023-02209-1 |
|
B Moal, A Orieux, T Ferté, A Neuraz, GA Brat, P Avillach, CL Bonzel, T Cai, K Cho, S Cossin, R Griffier, DA Hanauer, C Haverkamp, YL Ho, C Hong, MR Hutch, JG Klann, TT Le, NHW Loh, Y Luo, A Makoudjou, M Morris, DL Mowery, KL Olson, LP Patel, MJ Samayamuthu, FJ Sanz Vidorreta, ER Schriver, P Schubert, G Verdy, S Visweswaran, X Wang, GM Weber, Z Xia, W Yuan, HG Zhang, D Zöller, IS Kohane, The Consortium for Clinical Characterization of COVID-19 by EHR (4CE), A Boyer, V Jouhet. “Acute respiratory distress syndrome after SARS-CoV-2 infection on young adult population: International observational federated study based on electronic health records through the 4CE consortium”, PLOS ONE 18(1):e0266985 (2023). doi:10.1371/journal.pone.0266985 |

|
T Manz, S L’Yi, N Gehlenborg. “Gos: a declarative library for interactive genomics visualization in Python”, Bioinformatics 39(1):btad050 (2023). doi:10.1093/bioinformatics/btad050 |
|
S L’Yi, Q Wang, N Gehlenborg. “The Role of Visualization in Genomics Data Analysis Workflows: The Interview”, 2023 IEEE Visualization and Visual Analytics (VIS) (2023). doi:10.1109/VIS54172.2023.00029 |

|
S L’Yi, MS Keller, A Dandawate, L Taing, CH Chen, M Brown, CA Meyer, N Gehlenborg. “Cistrome Explorer: An Interactive Visual Analysis Tool for Large-Scale Epigenomic Data”, Bioinformatics 39(2):btad018 (2023). doi:10.1093/bioinformatics/btad018 |

|
S L’Yi, D Maziec, V Stevens, T Manz, A Veit, M Berselli, PJ Park, D Głodzik, N Gehlenborg. “Chromoscope: interactive multiscale visualization for structural variation in human genomes”, Nature Methods 20(12):1834-1835 (2023). doi:10.1038/s41592-023-02056-x |
|
S Jain, L Pei, JM Spraggins, M Angelo, JP Carson, N Gehlenborg, F Ginty, JP Gonçalves, JS Hagood, JW Hickey, NL Kelleher, LC Laurent, S Lin, Y Lin, H Liu, A Naba, ES Nakayasu, WJ Qian, A Radtke, P Robson, BR Stockwell, R Van De Plas, IS Vlachos, M Zhou, HuBMAP Consortium, KJ Ahn, J Allen, DM Anderson, CR Anderton, C Curcio, A Angelin, C Arvanitis, L Atta, D Awosika-Olumo, A Bahmani, H Bai, K Balderrama, L Balzano, G Bandyopadhyay, S Bandyopadhyay, Z Bar-Joseph, K Barnhart, D Barwinska, M Becich, L Becker, W Becker, K Bedi, S Bendall, K Benninger, D Betancur, K Bettinger, S Billings, P Blood, D Bolin, S Border, M Bosse, L Bramer, M Brewer, M Brusko, A Bueckle, K Burke, K Burnum-Johnson, E Butcher, E Butterworth, L Cai, R Calandrelli, M Caldwell, M Campbell-Thompson, D Cao, I Cao-Berg, R Caprioli, C Caraccio, A Caron, M Carroll, C Chadwick, A Chen, D Chen, F Chen, H Chen, J Chen, L Chen, L Chen, K Chiacchia, S Cho, P Chou, L Choy, C Cisar, G Clair, L Clarke, KA Clouthier, ME Colley, K Conlon, J Conroy, K Contrepois, A Corbett, A Corwin, D Cotter, E Courtois, A Cruz, C Csonka, K Czupil, V Daiya, K Dale, SA Davanagere, M Dayao, MP De Caestecker, A Decker, S Deems, D Degnan, T Desai, V Deshpande, G Deutsch, M Devlin, D Diep, C Dodd, S Donahue, W Dong, R Dos Santos Peixoto, M Duffy, M Dufresne, TE Duong, J Dutra, MT Eadon, TM El-Achkar, A Enninful, G Eraslan, D Eshelman, A Espin-Perez, ED Esplin, A Esselman, LD Falo, L Falo, J Fan, R Fan, MA Farrow, N Farzad, P Favaro, J Fermin, F Filiz, S Filus, K Fisch, E Fisher, S Fisher, K Flowers, WF Flynn, AB Fogo, D Fu, J Fulcher, A Fung, D Furst, M Gallant, F Gao, Y Gao, K Gaulton, JP Gaut, J Gee, RR Ghag, S Ghazanfar, S Ghose, D Gisch, I Gold, A Gondalia, B Gorman, W Greenleaf, N Greenwald, B Gregory, R Guo, R Gupta, H Hakimian, J Haltom, M Halushka, KS Han, C Hanson, P Harbury, J Hardi, L Harlan, RC Harris, A Hartman, E Heidari, J Helfer, D Helminiak, M Hemberg, N Henning, BW Herr, J Ho, J Holden-Wiltse, SH Hong, YK Hong, B Honick, G Hood, P Hu, Q Hu, M Huang, H Huyck, T Imtiaz, OG Isberg, M Itkin, D Jackson, M Jacobs, Y Jain, D Jewell, L Jiang, ZG Jiang, S Johnston, P Joshi, Y Ju, A Judd, A Kagel, A Kahn, N Kalavros, K Kalhor, D Karagkouni, T Karathanos, A Karunamurthy, S Katari, H Kates, M Kaushal, N Keener, M Keller, M Kenney, C Kern, P Kharchenko, J Kim, C Kingsford, J Kirwan, V Kiselev, J Kishi, RB Kitata, A Knoten, C Kollar, P Krishnamoorthy, ARS Kruse, K Da, A Kundaje, E Kutschera, Y Kwon, BB Lake, S Lancaster, J Langlieb, R Lardenoije, M Laronda, J Laskin, K Lau, H Lee, M Lee, M Lee, YL Strekalova, D Li, J Li, J Li, X Li, Z Li, YC Liao, T Liaw, P Lin, Y Lin, S Lindsay, C Liu, Y Liu, Y Liu, M Lott, M Lotz, L Lowery, P Lu, X Lu, N Lucarelli, X Lun, Z Luo, J Ma, E Macosko, M Mahajan, L Maier, D Makowski, M Malek, D Manthey, T Manz, K Margulies, J Marioni, M Martindale, C Mason, C Mathews, P Maye, C McCallum, E McDonough, L McDonough, H Mcdowell, M Meads, M Medina-Serpas, RM Ferreira, J Messinger, K Metis, LG Migas, B Miller, S Mimar, B Minor, R Misra, A Missarova, C Mistretta, R Moens, E Moerth, J Moffitt, G Molla, M Monroe, E Monte, M Morgan, D Muraro, B Murphy, E Murray, MA Musen, A Naglah, C Nasamran, T Neelakantan, S Nevins, H Nguyen, N Nguyen, T Nguyen, T Nguyen, D Nigra, M Nofal, G Nolan, G Nwanne, M O’Connor, K Okuda, M Olmer, K O’Neill, N Otaluka, M Pang, M Parast, L Pasa-Tolic, B Paten, NH Patterson, T Peng, G Phillips, M Pichavant, P Piehowski, H Pilner, E Pingry, Y Pita-Juarez, S Plevritis, A Ploumakis, A Pouch, G Pryhuber, J Puerto, D Qaurooni, L Qin, EM Quardokus, P Rajbhandari, R Rakow-Penner, R Ramasamy, D Read, EG Record, D Reeves, A Ricarte, A Rodríguez-Soto, A Ropelewski, J Rosario, MA Roselkis, D Rowe, TK Roy, M Ruffalo, N Ruschman, A Sabo, N Sachdev, S Saka, D Salamon, P Sarder, H Sasaki, R Satija, D Saunders, R Sawka, K Schey, H Schlehlein, D Scholten, S Schultz, L Schwartz, M Schwenk, R Scibek, A Segre, M Serrata, W Shands, X Shen, J Shendure, H Shephard, L Shi, T Shi, DG Shin, B Shirey, M Sibilla, M Silber, J Silverstein, D Simmel, A Simmons, D Singhal, S Sivajothi, T Smits, F Soncin, Q Song, V Stanley, T Stuart, H Su, P Su, X Sun, C Surrette, H Swahn, K Tan, S Teichmann, A Tejomay, G Tellides, K Thomas, T Thomas, M Thompson, H Tian, L Tideman, C Trapnell, AG Tsai, CF Tsai, L Tsai, E Tsui, T Tsui, J Tung, M Turner, J Uranic, ED Vaishnav, SR Varra, V Vaskivskyi, D Velickovic, M Velickovic, J Verheyden, J Waldrip, D Wallace, X Wan, A Wang, F Wang, M Wang, S Wang, X Wang, C Wasserfall, L Wayne, J Webber, GM Weber, B Wei, JJ Wei, A Weimer, J Welling, X Wen, Z Wen, MK Williams, S Winfree, N Winograd, A Woodard, D Wright, F Wu, PH Wu, Q Wu, X Wu, Y Xing, T Xu, M Yang, M Yang, J Yap, DH Ye, P Yin, Z Yuan, C Yun, A Zahraei, K Zemaitis, B Zhang, C Zhang, C Zhang, C Zhang, K Zhang, S Zhang, T Zhang, Y Zhang, B Zhao, W Zhao, JW Zheng, S Zhong, B Zhu, C Zhu, D Zhu, Q Zhu, Y Zhu, K Börner, MP Snyder. “Advances and prospects for the Human BioMolecular Atlas Program (HuBMAP)”, Nature Cell Biology 25(8):1089-1100 (2023). doi:10.1038/s41556-023-01194-w |

|
HG Zhang, JP Honerlaw, M Maripuri, MJ Samayamuthu, BR Beaulieu-Jones, HS Baig, S L’Yi, YL Ho, M Morris, VA Panickan, X Wang, GM Weber, KP Liao, S Visweswaran, BWQ Tan, W Yuan, N Gehlenborg, S Muralidhar, RB Ramoni, The Consortium for Clinical Characterization of COVID-19 by EHR (4CE), IS Kohane, Z Xia, K Cho, T Cai, GA Brat. “Potential pitfalls in the use of real world data to study Long COVID”, Nature Medicine 29(5):1040-1043 (2023). doi:10.1038/s41591-023-02274-y |
|
A Dagliati, ZH Strasser, ZS Hossein Abad, JG Klann, KB Wagholikar, R Mesa, S Visweswaran, M Morris, Y Luo, DW Henderson, MJ Samayamuthu, BWQ Tan, G Verdy, GS Omenn, Z Xia, R Bellazzi, JR Aaron, G Agapito, A Albayrak, G Albi, M Alessiani, A Alloni, DF Amendola, F Angoulvant, LLLJ Anthony, BJ Aronow, F Ashraf, A Atz, P Avillach, PS Azevedo, J Balshi, BK Beaulieu-Jones, DS Bell, A Bellasi, R Bellazzi, V Benoit, M Beraghi, JL Bernal-Sobrino, M Bernaux, R Bey, S Bhatnagar, A Blanco-Martínez, CL Bonzel, J Booth, S Bosari, FT Bourgeois, RL Bradford, GA Brat, S Bréant, NW Brown, R Bruno, WA Bryant, M Bucalo, E Bucholz, A Burgun, T Cai, M Cannataro, A Carmona, C Caucheteux, J Champ, J Chen, KY Chen, L Chiovato, L Chiudinelli, K Cho, JJ Cimino, TK Colicchio, S Cormont, S Cossin, JB Craig, JL Cruz-Bermúdez, J Cruz-Rojo, A Dagliati, M Daniar, C Daniel, P Das, B Devkota, A Dionne, R Duan, J Dubiel, SL DuVall, L Esteve, H Estiri, S Fan, RW Follett, T Ganslandt, NG Barrio, LX Garmire, N Gehlenborg, EJ Getzen, A Geva, T Gradinger, A Gramfort, R Griffier, N Griffon, O Grisel, A Gutiérrez-Sacristán, L Han, DA Hanauer, C Haverkamp, DY Hazard, B He, DW Henderson, M Hilka, YL Ho, JH Holmes, C Hong, KM Huling, MR Hutch, RW Issitt, AS Jannot, V Jouhet, R Kavuluru, MS Keller, CJ Kennedy, DA Key, K Kirchoff, JG Klann, IS Kohane, ID Krantz, D Kraska, AK Krishnamurthy, S L’Yi, TT Le, J Leblanc, G Lemaitre, L Lenert, D Leprovost, M Liu, NH Will Loh, Q Long, S Lozano-Zahonero, Y Luo, KE Lynch, S Mahmood, SE Maidlow, A Makoudjou, A Malovini, KD Mandl, C Mao, A Maram, P Martel, MR Martins, JS Marwaha, AJ Masino, M Mazzitelli, A Mensch, M Milano, MF Minicucci, B Moal, TM Ahooyi, JH Moore, C Moraleda, JS Morris, M Morris, KL Moshal, S Mousavi, DL Mowery, DA Murad, SN Murphy, TP Naughton, CT Breda Neto, A Neuraz, J Newburger, KY Ngiam, WFM Njoroge, JB Norman, J Obeid, MP Okoshi, KL Olson, GS Omenn, N Orlova, BD Ostasiewski, NP Palmer, N Paris, LP Patel, M Pedrera-Jiménez, ER Pfaff, AC Pfaff, D Pillion, S Pizzimenti, HU Prokosch, RA Prudente, A Prunotto, V Quirós-González, RB Ramoni, M Raskin, S Rieg, G Roig-Domínguez, P Rojo, P Rubio-Mayo, P Sacchi, C Sáez, E Salamanca, MJ Samayamuthu, LN Sanchez-Pinto, A Sandrin, N Santhanam, JCC Santos, FJ Sanz Vidorreta, M Savino, ER Schriver, P Schubert, J Schuettler, L Scudeller, NJ Sebire, P Serrano-Balazote, P Serre, A Serret-Larmande, M Shah, ZS Hossein Abad, D Silvio, P Sliz, J Son, C Sonday, AM South, A Spiridou, ZH Strasser, ALM Tan, BWQ Tan, BWL Tan, SE Tanni, DM Taylor, AI Terriza-Torres, V Tibollo, P Tippmann, EMS Toh, C Torti, EM Trecarichi, YJ Tseng, AK Vallejos, G Varoquaux, ME Vella, G Verdy, JJ Vie, S Visweswaran, M Vitacca, KB Wagholikar, LR Waitman, X Wang, D Wassermann, GM Weber, M Wolkewitz, S Wong, Z Xia, X Xiong, Y Ye, N Yehya, W Yuan, A Zambelli, HG Zhang, D Zo¨ller, V Zuccaro, C Zucco, SN Murphy, JH Holmes, H Estiri. “Characterization of long COVID temporal sub-phenotypes by distributed representation learning from electronic health record data: a cohort study”, eClinicalMedicine 64:102210 (2023). doi:10.1016/j.eclinm.2023.102210 |

|
Z Chen, C Zhang, Q Wang, J Troidl, S Warchol, J Beyer, N Gehlenborg, H Pfister. “Beyond Generating Code: Evaluating GPT on a Data Visualization Course”, 2023 IEEE VIS Workshop on Visualization Education, Literacy, and Activities (EduVis) (2023). doi:10.1109/EduVis60792.2023.00009 |
2022 | |

|
HG Zhang, A Dagliati, Z Shakeri Hossein Abad, X Xiong, CL Bonzel, Z Xia, BWQ Tan, P Avillach, GA Brat, C Hong, M Morris, S Visweswaran, LP Patel, A Gutiérrez-Sacristán, DA Hanauer, JH Holmes, MJ Samayamuthu, FT Bourgeois, S L’Yi, SE Maidlow, B Moal, SN Murphy, ZH Strasser, A Neuraz, KY Ngiam, NHW Loh, GS Omenn, A Prunotto, LA Dalvin, JG Klann, P Schubert, FJS Vidorreta, V Benoit, G Verdy, R Kavuluru, H Estiri, Y Luo, A Malovini, V Tibollo, R Bellazzi, K Cho, YL Ho, ALM Tan, BWL Tan, N Gehlenborg, S Lozano-Zahonero, V Jouhet, L Chiovato, BJ Aronow, EMS Toh, WGS Wong, S Pizzimenti, KB Wagholikar, M Bucalo, The Consortium for Clinical Characterization of COVID-19 by EHR (4CE), T Cai, AM South, IS Kohane, GM Weber. “International electronic health record-derived post-acute sequelae profiles of COVID-19 patients”, npj Digital Medicine 5(1):81 (2022). doi:10.1038/s41746-022-00623-8 |

|
F Cheng, MS Keller, H Qu, N Gehlenborg, Q Wang. “Polyphony: an Interactive Transfer Learning Framework for Single-Cell Data Analysis”, IEEE Transactions on Visualization and Computer Graphics 29(1):591-601 (2022). doi:10.1109/TVCG.2022.3209408
|
|
Q Wang, K Huang, P Chandak, M Zitnik, N Gehlenborg. “Extending the Nested Model for User-Centric XAI: A Design Study on GNN-based Drug Repurposing”, IEEE Transactions on Visualization and Computer Graphics 29(1):1266-1276 (2022). doi:10.1109/TVCG.2022.3209435
|
|
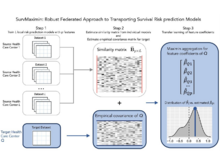
X Wang, HG Zhang, X Xiong, C Hong, GM Weber, GA Brat, CL Bonzel, Y Luo, R Duan, NP Palmer, MR Hutch, A Gutiérrez-Sacristán, R Bellazzi, L Chiovato, K Cho, A Dagliati, H Estiri, N García-Barrio, R Griffier, DA Hanauer, YL Ho, JH Holmes, MS Keller, JG Klann MEng, S L’Yi, S Lozano-Zahonero, SE Maidlow, A Makoudjou, A Malovini, B Moal, JH Moore, M Morris, DL Mowery, SN Murphy, A Neuraz, K Yuan Ngiam, GS Omenn, LP Patel, M Pedrera-Jiménez, A Prunotto, M Jebathilagam Samayamuthu, FJ Sanz Vidorreta, ER Schriver, P Schubert, P Serrano-Balazote, AM South, ALM Tan, BWL Tan, V Tibollo, P Tippmann, S Visweswaran, Z Xia, W Yuan, D Zöller, IS Kohane, P Avillach, Z Guo, T Cai, The Consortium for Clinical Characterization of COVID-19 by EHR (4CE). “SurvMaximin: Robust federated approach to transporting survival risk prediction models”, Journal of Biomedical Informatics 134:104176 (2022). doi:10.1016/j.jbi.2022.104176 |

|
SB Reiff, AJ Schroeder, K Kırlı, A Cosolo, C Bakker, L Mercado, S Lee, AD Veit, AK Balashov, C Vitzthum, W Ronchetti, KM Pitman, J Johnson, SR Ehmsen, P Kerpedjiev, N Abdennur, M Imakaev, SU Öztürk, U Çamoğlu, LA Mirny, N Gehlenborg, BH Alver, PJ Park. “The 4D Nucleome Data Portal as a resource for searching and visualizing curated nucleomics data”, Nature Communications 13(1):2365 (2022). doi:10.1038/s41467-022-29697-4 |
|
T Manz, I Gold, NH Patterson, C McCallum, MS Keller, BW Herr, K Börner, JM Spraggins, N Gehlenborg. “Viv: Multiscale Visualization of High-Resolution Multiplexed Bioimaging Data on the Web”, Nature Methods 19(5):515-516 (2022). doi:10.1038/s41592-022-01482-7 |
|
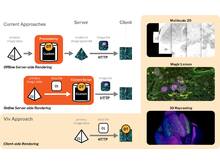
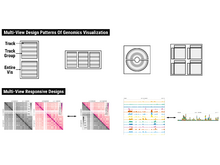
S L’Yi and N Gehlenborg. “Multi-View Design Patterns and Responsive Visualization for Genomics Data”, IEEE Transactions on Visualization and Computer Graphics 29(1):559-569 (2022). doi:10.1109/TVCG.2022.3209398 |
|
TS Liaw and N Gehlenborg. “User-Centric Process of Designing a Molecular & Cellular Query Interface for Biomedical Research”, Proceedings of the Design Society 2:221-230 (2022). doi:10.1017/pds.2022.24 |
|
MR Hutch, J Son, TT Le, C Hong, X Wang, Z Shakeri Hossein Abad, M Morris, A Gutiérrez-Sacristán, JG Klann, A Spiridou, R Bellazzi, V Benoit, CL Bonzel, WA Bryant, K Cho, P Das, DA Hanauer, DW Henderson, YL Ho, NHW Loh, A Makoudjou, A Malovini, B Moal, DL Mowery, MJ Samayamuthu, FJ Sanz Vidorreta, ER Schriver, P Schubert, J Talbert, AL Tan, BW Tan, BW Tan, V Tibollo, W Yuan, P Avillach, N Gehlenborg, GS Omenn, S Visweswaran, T Cai, Y Luo, Z Xia. “Neurological Diagnoses in Hospitalized COVID-19 Patients Associated With Adverse Outcomes: A Multinational Cohort Study”, SSRN Electronic Journal (2022). doi:10.2139/ssrn.4057133 |
|
|
C Hong, HG Zhang, S L’Yi, G Weber, P Avillach, BWQ Tan, A Gutiérrez-Sacristán, CL Bonzel, NP Palmer, A Malovini, V Tibollo, Y Luo, MR Hutch, M Liu, F Bourgeois, R Bellazzi, L Chiovato, FJ Sanz Vidorreta, TT Le, X Wang, W Yuan, A Neuraz, V Benoit, B Moal, M Morris, DA Hanauer, S Maidlow, K Wagholikar, S Murphy, H Estiri, A Makoudjou, P Tippmann, J Klann, RW Follett, N Gehlenborg, GS Omenn, Z Xia, A Dagliati, S Visweswaran, LP Patel, DL Mowery, ER Schriver, MJ Samayamuthu, R Kavuluru, S Lozano-Zahonero, D Zöller, ALM Tan, BWL Tan, KY Ngiam, JH Holmes, P Schubert, K Cho, YL Ho, BK Beaulieu-Jones, M Pedrera-Jiménez, N García-Barrio, P Serrano-Balazote, I Kohane, The Consortium for Clinical Characterization of COVID-19 by EHR (4CE), A South, GA Brat, T Cai. “Changes in laboratory value improvement and mortality rates over the course of the pandemic: an international retrospective cohort study of hospitalised patients infected with SARS-CoV-2”, BMJ Open 12(6):e057725 (2022). doi:10.1136/bmjopen-2021-057725 |

|
A Gutiérrez-Sacristán, A Serret-Larmande, MR Hutch, C Sáez, BJ Aronow, S Bhatnagar, CL Bonzel, T Cai, B Devkota, DA Hanauer, NHW Loh, Y Luo, B Moal, TM Ahooyi, WFM Njoroge, GS Omenn, LN Sanchez-Pinto, AM South, F Sperotto, ALM Tan, DM Taylor, G Verdy, S Visweswaran, Z Xia, J Zahner, P Avillach, FT Bourgeois, Consortium for Clinical Characterization of COVID-19 by EHR (4CE), JR Aaron, G Agapito, A Albayrak, G Albi, M Alessiani, A Alloni, DF Amendola, F Angoulvant, LLLJ Anthony, F Ashraf, A Atz, PS Azevedo, J Balshi, BK Beaulieu-Jones, DS Bell, A Bellasi, R Bellazzi, V Benoit, M Beraghi, JL Bernal-Sobrino, M Bernaux, R Bey, A Blanco-Martínez, M Boeker, J Booth, S Bosari, RL Bradford, GA Brat, S Bréant, NW Brown, R Bruno, WA Bryant, M Bucalo, E Bucholz, A Burgun, M Cannataro, A Carmona, C Caucheteux, J Champ, J Chen, KY Chen, L Chiovato, L Chiudinelli, K Cho, JJ Cimino, TK Colicchio, S Cormont, S Cossin, JB Craig, JL Cruz-Bermúdez, J Cruz-Rojo, A Dagliati, M Daniar, C Daniel, P Das, A Dionne, R Duan, J Dubiel, SL DuVall, L Esteve, H Estiri, S Fan, RW Follett, T Ganslandt, N García-Barrio, LX Garmire, N Gehlenborg, EJ Getzen, A Geva, T González González, T Gradinger, A Gramfort, R Griffier, N Griffon, O Grisel, PH Guzzi, L Han, C Haverkamp, DY Hazard, B He, DW Henderson, M Hilka, YL Ho, JH Holmes, C Hong, KM Huling, RW Issitt, AS Jannot, V Jouhet, R Kavuluru, MS Keller, CJ Kennedy, KF Kernan, DA Key, K Kirchoff, JG Klann, IS Kohane, ID Krantz, D Kraska, AK Krishnamurthy, S L’Yi, TT Le, J Leblanc, G Lemaitre, L Lenert, D Leprovost, M Liu, Q Long, S Lozano-Zahonero, KE Lynch, S Mahmood, SE Maidlow, A Makoudjou, A Malovini, KD Mandl, C Mao, A Maram, P Martel, MR Martins, JS Marwaha, AJ Masino, M Mazzitelli, A Mensch, M Milano, MF Minicucci, JH Moore, C Moraleda, JS Morris, M Morris, KL Moshal, S Mousavi, DL Mowery, DA Murad, SN Murphy, TP Naughton, CTB Neto, A Neuraz, J Newburger, KY Ngiam, JB Norman, J Obeid, MP Okoshi, KL Olson, N Orlova, BD Ostasiewski, NP Palmer, N Paris, LP Patel, M Pedrera-Jiménez, AC Pfaff, ER Pfaff, D Pillion, S Pizzimenti, T Priya, HU Prokosch, RA Prudente, A Prunotto, V Quirós-González, RB Ramoni, M Raskin, S Rieg, G Roig-Domínguez, P Rojo, P Rubio-Mayo, P Sacchi, E Salamanca, MJ Samayamuthu, A Sandrin, N Santhanam, JC Santos, FJ Sanz Vidorreta, M Savino, ER Schriver, P Schubert, J Schuettler, L Scudeller, NJ Sebire, P Serrano-Balazote, P Serre, M Shah, Z Shakeri Hossein Abad, D Silvio, P Sliz, J Son, C Sonday, A Spiridou, ZH Strasser, BW Tan, BW Tan, SE Tanni, AI Terriza-Torres, V Tibollo, P Tippmann, EM Toh, C Torti, EM Trecarichi, AK Vallejos, G Varoquaux, ME Vella, JJ Vie, M Vitacca, KB Wagholikar, LR Waitman, X Wang, D Wassermann, GM Weber, M Wolkewitz, S Wong, X Xiong, Y Ye, N Yehya, W Yuan, A Zambelli, HG Zhang, D Zöller, V Zuccaro, C Zucco. “Hospitalizations Associated With Mental Health Conditions Among Adolescents in the US and France During the COVID-19 Pandemic”, JAMA Network Open 5(12):e2246548 (2022). doi:10.1001/jamanetworkopen.2022.46548 |

|
U Gisladottir, D Nakikj, R Jhunjhunwala, J Panton, G Brat, N Gehlenborg. “Effective Communication of Personalized Risks and Patient Preferences During Surgical Informed Consent Using Data Visualization: Qualitative Semistructured Interview Study With Patients After Surgery”, JMIR Human Factors 9(2):e29118 (2022). doi:10.2196/29118 |
|
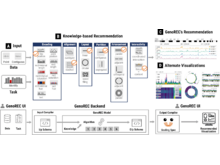
A Pandey, S L’Yi, Q Wang, MA Borkin, N Gehlenborg. “GenoREC: A Recommendation System for Interactive Genomics Data Visualization”, IEEE Transactions on Visualization and Computer Graphics 29(1):570-580 (2022). doi:10.1109/TVCG.2022.3209407 |
|
GM Weber, C Hong, Z Xia, NP Palmer, P Avillach, S L’Yi, MS Keller, SN Murphy, A Gutiérrez-Sacristán, CL Bonzel, A Serret-Larmande, A Neuraz, GS Omenn, S Visweswaran, JG Klann, AM South, NHW Loh, M Cannataro, BK Beaulieu-Jones, R Bellazzi, G Agapito, M Alessiani, BJ Aronow, DS Bell, V Benoit, FT Bourgeois, L Chiovato, K Cho, A Dagliati, SL DuVall, NG Barrio, DA Hanauer, YL Ho, JH Holmes, RW Issitt, M Liu, Y Luo, KE Lynch, SE Maidlow, A Malovini, KD Mandl, C Mao, ME Matheny, JH Moore, JS Morris, M Morris, DL Mowery, KY Ngiam, LP Patel, M Pedrera-Jimenez, RB Ramoni, ER Schriver, P Schubert, PS Balazote, A Spiridou, ALM Tan, BWL Tan, V Tibollo, C Torti, EM Trecarichi, X Wang, The Consortium for Clinical Characterization of COVID-19 by EHR (4CE) (incl. N Gehlenborg), IS Kohane, T Cai, GA Brat. “International comparisons of laboratory values from the 4CE collaborative to predict COVID-19 mortality”, npj Digital Medicine 5(1):74 (2022). doi:10.1038/s41746-022-00601-0 |

|
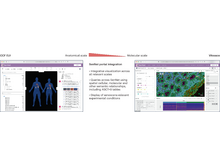
MR Hutch, J Son, TT Le, C Hong, X Wang, Z Shakeri Hossein Abad, M Morris, A Gutiérrez-Sacristán, JG Klann, A Spiridou, A Batugo, R Bellazzi, V Benoit, CL Bonzel, WA Bryant, L Chiudinelli, K Cho, P Das, T González González, DA Hanauer, DW Henderson, YL Ho, NHW Loh, A Makoudjou, S Makwana, A Malovini, B Moal, DL Mowery, A Neuraz, MJ Samayamuthu, FJ Sanz Vidorreta, ER Schriver, P Schubert, J Talbert, ALM Tan, BWL Tan, BWQ Tan, V Tibollo, P Tippman, G Verdy, W Yuan, P Avillach, N Gehlenborg, GS Omenn, The Consortium for Clinical Characterization of COVID-19 by EHR (4CE), S Visweswaran, T Cai, Y Luo, Z Xia. “Neurological diagnoses in hospitalized COVID-19 patients associated with adverse outcomes: a multinational cohort stud”, PLOS Digital Health 3(4):e0000484 (2022). doi:10.1371/journal.pdig.0000484 |
|
SenNet Consortium, Writing Group, PJ Lee, CC Benz, P Blood, K Börner, J Campisi, F Chen, H Daldrup-Link, P De Jager, L Ding, FE Duncan, O Eickelberg, R Fan, T Finkel, D Furman, V Garovic, N Gehlenborg, C Glass, I Heckenbach, ZB Joseph, P Katiyar, SJ Kim, M Königshoff, GA Kuchel, H Lee, JH Lee, J Ma, Q Ma, S Melov, K Metis, AL Mora, N Musi, N Neretti, JF Passos, I Rahman, JC Rivera-Mulia, P Robson, M Rojas, AL Roy, M Scheibye-Knudsen, B Schilling, P Shi, JC Silverstein, V Suryadevara, J Xie, J Wang, AI Wong, LJ Niedernhofer, Brown University TDA, S Wang, Buck Institute for Research on Aging TMC/TDA, H Anvari, J Balough, C Benz, J Bons, B Brenerman, W Evans, A Gerencser, H Gregory, M Hansen, J Justice, P Kapahi, N Murad, A O’Broin, ME Pavone, M Powell, G Scott, E Shanes, M Shankaran, E Verdin, D Winer, F Wu, Consortium Organization and Data Coordinating Center (CODCC), A Adams, PD Blood, A Bueckle, I Cao-Berg, H Chen, M Davis, S Filus, Y Hao, A Hartman, E Hasanaj, J Helfer, B Herr, ZB Joseph, G Molla, G Mou, J Puerto, EM Quardokus, AJ Ropelewski, M Ruffalo, R Satija, M Schwenk, R Scibek, W Shirey, M Sibilla, J Welling, Z Yuan, Columbia TMC, R Bonneau, A Christiano, B Izar, V Menon, DM Owens, H Phatnani, C Smith, Y Suh, AF Teich, Duke University TMC, V Bekker, C Chan, E Coutavas, MG Hartwig, Z Ji, AB Nixon, Massachusetts General Hospital TDA, Z Dou, J Rajagopal, N Slavov, Mayo Clinic TDA, D Holmes, D Jurk, JL Kirkland, A Lagnado, T Tchkonia, National Institute of Health (NIH), K Abraham, A Dibattista, YW Fridell, TK Howcroft, C Jhappan, VP Montes, M Prabhudas, H Resat, V Taylor, Stanford TDA, M Kumar, V Suryadevara, University of Connecticut TMC, F Cigarroa, R Cohn, TM Cortes, E Courtois, J Chuang, M Davé, S Domanskyi, EAL Enninga, GN Eryilmaz, SE Espinoza, J Gelfond, J Kirkland, GA Kuchel, CL Kuo, JS Lehman, C Aguayo-Mazzucato, A Meves, M Rani, S Sanders, A Thibodeau, SG Tullius, D Ucar, B White, Q Wu, M Xu, S Yamaguchi, University of Michigan TDA, N Assarzadegan, CS Cho, I Hwang, Y Hwang, J Xi, University of Minnesota TMC, OA Adeyi, CF Aliferis, A Bartolomucci, X Dong, MJ DuFresne-To, S Ikramuddin, SG Johnson, AC Nelson, LJ Niedernhofer, XS Revelo, C Trevilla-Garcia, JM Sedivy, EL Thompson, PD Robbins, J Wang, University of Pittsburgh TMC, KM Aird, JK Alder, D Beaulieu, M Bueno, J Calyeca, JA Chamucero-Millaris, SY Chan, D Chung, A Corbett, V Gorbunova, KM Gowdy, A Gurkar, JC Horowitz, Q Hu, G Kaur, TO Khaliullin, R Lafyatis, S Lanna, D Li, A Ma, A Morris, TM Muthumalage, V Peters, GS Pryhuber, BF Reader, L Rosas, JC Sembrat, S Shaikh, H Shi, SD Stacey, CS Croix, C Wang, Q Wang, A Watts, University of Washington TDA, L Gu, Y Lin, PS Rabinovitch, MT Sweetwyne, Washington University TMC, MN Artyomov, SJ Ballentine, MG Chheda, SR Davies, JF DiPersio, RC Fields, JAJ Fitzpatrick, RS Fulton, S Imai, S Jain, T Ju, VM Kushnir, DC Link, M Ben Major, ST Oh, D Rapp, MP Rettig, SA Stewart, DJ Veis, KR Vij, MC Wendl, MA Wyczalkowski, Yale TMC, JE Craft, A Enninful, N Farzad, P Gershkovich, S Halene, Y Kluger, J VanOudenhove, M Xu, J Yang, M Yang. “NIH SenNet Consortium to map senescent cells throughout the human lifespan to understand physiological health”, Nature Aging 2(12):1090-1100 (2022). doi:10.1038/s43587-022-00326-5 |
2021 | |

|
GM Weber, HG Zhang, S L’Yi, CL Bonzel, C Hong, P Avillach, A Gutiérrez-Sacristán, NP Palmer, ALM Tan, X Wang, W Yuan, N Gehlenborg, A Alloni, DF Amendola, A Bellasi, R Bellazzi, M Beraghi, M Bucalo, L Chiovato, K Cho, A Dagliati, H Estiri, RW Follett, N García Barrio, DA Hanauer, DW Henderson, YL Ho, JH Holmes, MR Hutch, R Kavuluru, K Kirchoff, JG Klann, AK Krishnamurthy, TT Le, M Liu, NHW Loh, S Lozano-Zahonero, Y Luo, S Maidlow, A Makoudjou, A Malovini, MR Martins, B Moal, M Morris, DL Mowery, SN Murphy, A Neuraz, KY Ngiam, MP Okoshi, GS Omenn, LP Patel, M Pedrera Jiménez, RA Prudente, MJ Samayamuthu, FJ Sanz Vidorreta, ER Schriver, P Schubert, P Serrano Balazote, BW Tan, SE Tanni, V Tibollo, S Visweswaran, KB Wagholikar, Z Xia, D Zöller, The Consortium for Clinical Characterization of COVID-19 by EHR (4CE), IS Kohane, T Cai, AM South, GA Brat. “Authorship Correction: International Changes in COVID-19 Clinical Trajectories Across 315 Hospitals and 6 Countries: Retrospective Cohort Study”, Journal of Medical Internet Research 23(11):e34625 (2021). doi:10.2196/34625 |

|
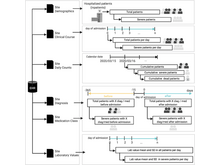
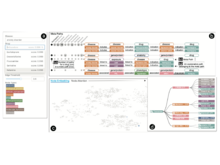
Q Wang, T Mazor, TA Harbig, E Cerami, N Gehlenborg. “ThreadStates: State-based Visual Analysis of Disease Progression”, IEEE Transactions on Visualization and Computer Graphics 28(1):238-247 (2021). doi:10.1109/TVCG.2021.3114840 |
|
Q Wang, K Huang, P Chandak, N Gehlenborg, M Zitnik. “Interactive Visual Explanations for Deep Drug Repurposing”, Workshop on Interpretable ML in Healthcare at International Conference on Machine Learning (ICML) (2021). |

|
S L’Yi, Q Wang, F Lekschas, N Gehlenborg. “Gosling: A Grammar-based Toolkit for Scalable and Interactive Genomics Data Visualization”, IEEE Transactions on Visualization and Computer Graphics 28(1):140-150 (2021). doi:10.1109/TVCG.2021.3114876 |
|
TT Le, A Gutiérrez-Sacristán, J Son, C Hong, AM South, BK Beaulieu-Jones, NHW Loh, Y Luo, M Morris, KY Ngiam, LP Patel, MJ Samayamuthu, E Schriver, ALM Tan, J Moore, T Cai, GS Omenn, P Avillach, IS Kohane, The Consortium for Clinical Characterization of COVID-19 by EHR (4CE), JR Aaron, G Agapito, A Albayrak, M Alessiani, DF Amendola, F Angoulvant, LLLJ Anthony, BJ Aronow, A Atz, J Balshi, DS Bell, A Bellasi, R Bellazzi, V Benoit, M Beraghi, JL Bernal Sobrino, M Bernaux, R Bey, A Blanco Martínez, M Boeker, CL Bonzel, J Booth, S Bosari, FT Bourgeois, RL Bradford, GA Brat, S Bréant, NW Brown, WA Bryant, M Bucalo, A Burgun, M Cannataro, A Carmona, C Caucheteux, J Champ, K Chen, J Chen, L Chiovato, L Chiudinelli, JJ Cimino, TK Colicchio, S Cormont, S Cossin, JB Craig, JL Cruz Bermúdez, J Cruz Rojo, A Dagliati, M Daniar, C Daniel, A Davoudi, B Devkota, J Dubiel, L Esteve, S Fan, RW Follett, PSA Gaiolla, T Ganslandt, N García Barrio, LX Garmire, N Gehlenborg, A Geva, T Gradinger, A Gramfort, R Griffier, N Griffon, O Grisel, DA Hanauer, C Haverkamp, B He, DW Henderson, M Hilka, JH Holmes, P Horki, KM Huling, MR Hutch, RW Issitt, AS Jannot, V Jouhet, R Kavuluru, MS Keller, K Kirchoff, JG Klann, ID Krantz, D Kraska, AK Krishnamurthy, S L’Yi, J Leblanc, ARR Leite, G Lemaitre, L Lenert, D Leprovost, M Liu, S Lozano-Zahonero, KE Lynch, S Mahmood, S Maidlow, AC Makoudjou Tchendjou, A Malovini, KD Mandl, C Mao, A Maram, P Martel, AJ Masino, ME Matheny, T Maulhardt, M Mazzitelli, MT McDuffie, A Mensch, F Ashraf, M Milano, MF Minicucci, B Moal, C Moraleda, JS Morris, KL Moshal, S Mousavi, DA Murad, SN Murphy, TP Naughton, A Neuraz, JB Norman, J Obeid, MP Okoshi, KL Olson, N Orlova, BD Ostasiewski, NP Palmer, N Paris, M Pedrera Jimenez, ER Pfaff, D Pillion, HU Prokosch, RA Prudente, V Quirós González, RB Ramoni, M Raskin, S Rieg, G Roig Domínguez, P Rojo, C Sáez, E Salamanca, A Sandrin, JCC Santos, M Savino, J Schuettler, L Scudeller, NJ Sebire, PS Balazote, P Serre, A Serret-Larmande, Z Shakeri, D Silvio, P Sliz, C Sonday, A Spiridou, BWQ Tan, BWL Tan, SE Tanni, DM Taylor, AI Terriza-Torres, V Tibollo, P Tippmann, C Torti, EM Trecarichi, YJ Tseng, AK Vallejos, G Varoquaux, M Vella, JJ Vie, M Vitacca, KB Wagholikar, LR Waitman, D Wassermann, GM Weber, Y William, N Yehya, A Zambelli, HG Zhang, D Zoeller, C Zucco, S Visweswaran, DL Mowery, Z Xia. “Multinational characterization of neurological phenotypes in patients hospitalized with COVID-19”, Scientific Reports 11(1):20238 (2021). doi:10.1038/s41598-021-99481-9 |
|
IS Kohane, BJ Aronow, P Avillach, BK Beaulieu-Jones, R Bellazzi, RL Bradford, GA Brat, M Cannataro, JJ Cimino, N García-Barrio, N Gehlenborg, M Ghassemi, A Gutiérrez-Sacristán, DA Hanauer, JH Holmes, C Hong, JG Klann, NHW Loh, Y Luo, KD Mandl, M Daniar, JH Moore, SN Murphy, A Neuraz, KY Ngiam, GS Omenn, N Palmer, LP Patel, M Pedrera-Jiménez, P Sliz, AM South, ALM Tan, DM Taylor, BW Taylor, C Torti, AK Vallejos, KB Wagholikar, The Consortium For Clinical Characterization Of COVID-19 By EHR (4CE), GM Weber, T Cai. “What Every Reader Should Know About Studies Using Electronic Health Record Data but May Be Afraid to Ask”, Journal of Medical Internet Research 23(3):e22219 (2021). doi:10.2196/22219 |
|
|
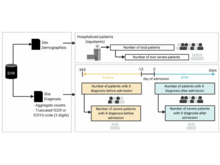
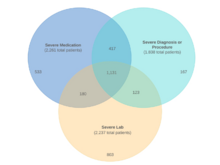
JG Klann, H Estiri, GM Weber, B Moal, P Avillach, C Hong, ALM Tan, BK Beaulieu-Jones, V Castro, T Maulhardt, A Geva, A Malovini, AM South, S Visweswaran, M Morris, MJ Samayamuthu, GS Omenn, KY Ngiam, KD Mandl, M Boeker, KL Olson, DL Mowery, RW Follett, DA Hanauer, R Bellazzi, JH Moore, NHW Loh, DS Bell, KB Wagholikar, L Chiovato, V Tibollo, S Rieg, ALLJ Li, V Jouhet, E Schriver, Z Xia, M Hutch, Y Luo, IS Kohane, The Consortium for Clinical Characterization of COVID-19 by EHR (4CE) (CONSORTIA AUTHOR), GA Brat, SN Murphy. “Validation of an internationally derived patient severity phenotype to support COVID-19 analytics from electronic health record data”, Journal of the American Medical Informatics Association 28(7):1411-1420 (2021). doi:10.1093/jamia/ocab018 |
|
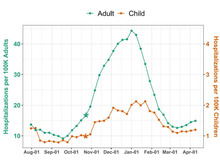
MR Hutch, M Liu, P Avillach, Consortium for Clinical Characterization of COVID-19 by EHR (4CE), Y Luo, FT Bourgeois. “National Trends in Disease Activity for COVID-19 Among Children in the US”, Frontiers in Pediatrics 9:700656 (2021). doi:10.3389/fped.2021.700656 |
|
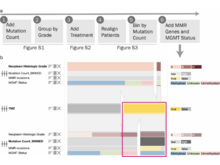
TA Harbig, S Nusrat, T Mazor, Q Wang, A Thomson, H Bitter, E Cerami, N Gehlenborg. “OncoThreads: Visualization of Large Scale Longitudinal Cancer Molecular Data”, Bioinformatics (Oxford, England) 37(Suppl_1):i59-i66 (2021). doi:10.1093/bioinformatics/btab289 |

|
GM Weber, HG Zhang, S L’Yi, CL Bonzel, C Hong, P Avillach, A Gutiérrez-Sacristán, NP Palmer, ALM Tan, X Wang, W Yuan, N Gehlenborg, A Alloni, DF Amendola, A Bellasi, R Bellazzi, M Beraghi, M Bucalo, L Chiovato, K Cho, A Dagliati, H Estiri, RW Follett, N García Barrio, DA Hanauer, DW Henderson, YL Ho, JH Holmes, MR Hutch, R Kavuluru, K Kirchoff, JG Klann, AK Krishnamurthy, TT Le, M Liu, NHW Loh, S Lozano-Zahonero, Y Luo, S Maidlow, A Makoudjou, A Malovini, MR Martins, B Moal, M Morris, DL Mowery, SN Murphy, A Neuraz, KY Ngiam, MP Okoshi, GS Omenn, LP Patel, M Pedrera Jiménez, RA Prudente, MJ Samayamuthu, FJ Sanz Vidorreta, ER Schriver, P Schubert, P Serrano Balazote, BW Tan, SE Tanni, V Tibollo, S Visweswaran, KB Wagholikar, Z Xia, D Zöller, The Consortium For Clinical Characterization Of COVID-19 By EHR (4CE), IS Kohane, T Cai, AM South, GA Brat. “International Changes in COVID-19 Clinical Trajectories Across 315 Hospitals and 6 Countries: a 4CE Consortium Study”, Journal of Medical Internet Research 23(10):e31400 (2021). doi:10.2196/31400 |
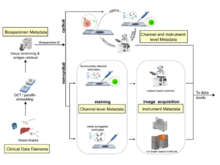
|
D Schapiro, C Yapp, A Sokolov, SM Reynolds, YA Chen, D Sudar, Y Xie, J Muhlich, R Arias-Camison, S Arena, AJ Taylor, M Nikolov, M Tyler, JR Lin, EA Burlingame, Human Tumor Atlas Network, YH Chang, SL Farhi, V Thorsson, N Venkatamohan, JL Drewes, D Pe’er, DA Gutman, MD Herrmann, N Gehlenborg, P Bankhead, JT Roland, JM Herndon, MP Snyder, M Angelo, G Nolan, JR Swedlow, N Schultz, DT Merrick, SA Mazzili, E Cerami, SJ Rodig, S Santagata, PK Sorger. “MITI Minimum Information guidelines for highly multiplexed tissue images”, Nature Methods 19(3):262-267 (2021). doi:10.1038/s41592-022-01415-4 |
|
The Consortium for Characterization of COVID-19 by EHR (4CE) (incl. M Keller, S L’Yi N Gehlenborg). “Evolving phenotypes of non-hospitalized patients that indicate long COVID”, BMC Medicine 19(1):249 (2021). doi:10.1186/s12916-021-02115-0 |

|
FT Bourgeois, A Gutiérrez-Sacristán, MS Keller, M Liu, C Hong, CL Bonzel, ALM Tan, BJ Aronow, M Boeker, J Booth, J Cruz-Rojo, B Devkota, N García-Barrio, N Gehlenborg, A Geva, DA Hanauer, MR Hutch, RW Issitt, JG Klann, Y Luo, KD Mandl, C Mao, B Moal, KL Moshal, SN Murphy, A Neuraz, KY Ngiam, GS Omenn, LP Patel, M Pedrera-Jiménez, NJ Sebire, P Serrano-Balazote, A Serret-Larmande, AM South, A Spiridou, DM Taylor, P Tippmann, S Visweswaran, GM Weber, IS Kohane, T Cai, P Avillach, Consortium for Clinical Characterization of COVID-19 by EHR (4CE). “International Analysis of Electronic Health Records of Children and Youth Hospitalized With COVID-19 Infection in 6 Countries”, JAMA Network Open 4(6):e2112596 (2021). doi:10.1001/jamanetworkopen.2021.12596 |
2020 | |

|
F Lekschas, X Zhou, W Chen, N Gehlenborg, B Bach, H Pfister. “A Generic Framework and Library for Exploration of Small Multiples through Interactive Piling”, IEEE Transactions on Visualization and Computer Graphics 27(2):358-368 (2020). doi:10.1109/TVCG.2020.3028948 |

|
F Lekschas, M Behrisch, B Bach, P Kerpedjiev, N Gehlenborg, H Pfister. “Pattern-Driven Navigation in 2D Multiscale Visualizations with Scalable Insets”, IEEE Transactions on Visualization and Computer Graphics 26(1):611-621 (2020). doi:10.1109/TVCG.2019.2934555 |
|
F Lekschas, B Peterson, D Haehn, E Ma, N Gehlenborg, H Pfister. “Peax: Interactive Visual Pattern Search in Sequential Data Using Unsupervised Deep Representation Learning”, Computer Graphics Forum 39(3):167-179 (2020). doi:10.1111/cgf.13971 |
|
M Goldman and N Gehlenborg. “Making Tools that People Will Use: User-Centered Design in Computational Biology Research”, Biocomputing 2021 (2020). doi:10.1142/9789811232701_0034 |
|
|
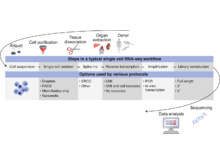
A Füllgrabe, N George, M Green, P Nejad, B Aronow, SK Fexova, C Fischer, MA Freeberg, L Huerta, N Morrison, RH Scheuermann, D Taylor, N Vasilevsky, L Clarke, N Gehlenborg, J Kent, J Marioni, S Teichmann, A Brazma, I Papatheodorou. “Guidelines for reporting single-cell RNA-seq experiments”, Nature Biotechnology 38(12):1384-1386 (2020). doi:10.1038/s41587-020-00744-z |
|
GA Brat, GM Weber, N Gehlenborg, P Avillach, NP Palmer, L Chiovato, J Cimino, LR Waitman, GS Omenn, A Malovini, JH Moore, BK Beaulieu-Jones, V Tibollo, SN Murphy, SL Yi, MS Keller, R Bellazzi, DA Hanauer, A Serret-Larmande, A Gutierrez-Sacristan, JJ Holmes, DS Bell, KD Mandl, RW Follett, JG Klann, DA Murad, L Scudeller, M Bucalo, K Kirchoff, J Craig, J Obeid, V Jouhet, R Griffier, S Cossin, B Moal, LP Patel, A Bellasi, HU Prokosch, D Kraska, P Sliz, ALM Tan, KY Ngiam, A Zambelli, DL Mowery, E Schiver, B Devkota, RL Bradford, M Daniar, C Daniel, V Benoit, R Bey, N Paris, P Serre, N Orlova, J Dubiel, M Hilka, AS Jannot, S Breant, J Leblanc, N Griffon, A Burgun, M Bernaux, A Sandrin, E Salamanca, S Cormont, T Ganslandt, T Gradinger, J Champ, M Boeker, P Martel, L Esteve, A Gramfort, O Grisel, D Leprovost, T Moreau, G Varoquaux, JJ Vie, D Wassermann, A Mensch, C Caucheteux, C Haverkamp, G Lemaitre, S Bosari, ID Krantz, A South, T Cai, IS Kohane. “International Electronic Health Record-Derived COVID-19 Clinical Course Profiles: the 4CE Consortium”, npj Digital Medicine 3(1):109 (2020). doi:10.1038/s41746-020-00308-0 |
2019 | |

|
S Nusrat, T Harbig, N Gehlenborg. “Tasks, Techniques, and Tools for Genomic Data Visualization”, Computer Graphics Forum 38(3):781-805 (2019). doi:10.1111/cgf.13727 |

|
B Morrow, T Manz, AE Chung, N Gehlenborg, D Gotz. “Periphery Plots for Contextualizing Heterogeneous Time-Based Charts”, 2019 IEEE Visualization Conference (VIS) (2019). doi:10.1109/VISUAL.2019.8933582 |
|
S Mataraso, V Socrates, F Lekschas, N Gehlenborg. “Halyos: A patient-facing visual EHR interface for longitudinal risk awareness”, ACI Open 06(02):e123-e128 (2019). doi:10.1055/s-0042-1749191 |
|
MP Snyder, S Lin, A Posgai, M Atkinson, A Regev, J Rood, O Rosen, L Gaffney, A Hupalowska, R Satija, N Gehlenborg, J Shendure, J Laskin, P Harbury, NA Nystrom, Z Bar-Joseph, K Zhang, K Borner, Y Lin, R Conroy, D Procaccini, AL Roy, A Pillai, M Brown, ZS Galis (for the HuBMAP Consortium). “The human body at cellular resolution: the NIH Human Biomolecular Atlas Program”, Nature 574(7777):187-192 (2019). doi:10.1038/s41586-019-1629-x |

|
K Gadhave, H Strobelt, N Gehlenborg, A Lex. “Upset 2: From prototype to tool”, Proceedings of the IEEE Information Visualization Conference–Posters (InfoVis’ 19) (2019). |

|
AE Chung, K Glass, J Leisey-Bartsch, L Mentch, N Gehlenborg, D Gotz. “Precision VISSTA: Bring-Your-Own-Device (BYOD) mHealth Data for Precision Health.”, AMIA (2019). |
2018 | |
|
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “Machine Learning Detects Pan-cancer Ras Pathway Activation in The Cancer Genome Atlas”, Cell Reports 23(1):172-180.e3 (2018). doi:10.1016/j.celrep.2018.03.046 |

|
Y Wang, X Xu, D Maglic, MT Dill, K Mojumdar, PKS Ng, KJ Jeong, YH Tsang, D Moreno, VH Bhavana, X Peng, Z Ge, H Chen, J Li, Z Chen, H Zhang, L Han, D Du, CJ Creighton, GB Mills, F Camargo, H Liang, SJ Caesar-Johnson, JA Demchok, I Felau, M Kasapi, ML Ferguson, CM Hutter, HJ Sofia, R Tarnuzzer, Z Wang, L Yang, JC Zenklusen, JJ Zhang, S Chudamani, J Liu, L Lolla, R Naresh, T Pihl, Q Sun, Y Wan, Y Wu, J Cho, T DeFreitas, S Frazer, N Gehlenborg, G Getz, DI Heiman, J Kim, MS Lawrence, P Lin, S Meier, MS Noble, G Saksena, D Voet, H Zhang, B Bernard, N Chambwe, V Dhankani, T Knijnenburg, R Kramer, K Leinonen, Y Liu, M Miller, S Reynolds, I Shmulevich, V Thorsson, W Zhang, R Akbani, BM Broom, AM Hegde, Z Ju, RS Kanchi, A Korkut, J Li, H Liang, S Ling, W Liu, Y Lu, GB Mills, KS Ng, A Rao, M Ryan, J Wang, JN Weinstein, J Zhang, A Abeshouse, J Armenia, D Chakravarty, WK Chatila, I De Bruijn, J Gao, BE Gross, ZJ Heins, R Kundra, K La, M Ladanyi, A Luna, MG Nissan, A Ochoa, SM Phillips, E Reznik, F Sanchez-Vega, C Sander, N Schultz, R Sheridan, SO Sumer, Y Sun, BS Taylor, J Wang, H Zhang, P Anur, M Peto, P Spellman, C Benz, JM Stuart, CK Wong, C Yau, DN Hayes, JS Parker, MD Wilkerson, A Ally, M Balasundaram, R Bowlby, D Brooks, R Carlsen, E Chuah, N Dhalla, R Holt, SJM Jones, K Kasaian, D Lee, Y Ma, MA Marra, M Mayo, RA Moore, AJ Mungall, K Mungall, AG Robertson, S Sadeghi, JE Schein, P Sipahimalani, A Tam, N Thiessen, K Tse, T Wong, AC Berger, R Beroukhim, AD Cherniack, C Cibulskis, SB Gabriel, GF Gao, G Ha, M Meyerson, SE Schumacher, J Shih, MH Kucherlapati, RS Kucherlapati, S Baylin, L Cope, L Danilova, MS Bootwalla, PH Lai, DT Maglinte, DJ Van Den Berg, DJ Weisenberger, JT Auman, S Balu, T Bodenheimer, C Fan, KA Hoadley, AP Hoyle, SR Jefferys, CD Jones, S Meng, PA Mieczkowski, LE Mose, AH Perou, CM Perou, J Roach, Y Shi, JV Simons, T Skelly, MG Soloway, D Tan, U Veluvolu, H Fan, T Hinoue, PW Laird, H Shen, W Zhou, M Bellair, K Chang, K Covington, CJ Creighton, H Dinh, HV Doddapaneni, LA Donehower, J Drummond, RA Gibbs, R Glenn, W Hale, Y Han, J Hu, V Korchina, S Lee, L Lewis, W Li, X Liu, M Morgan, D Morton, D Muzny, J Santibanez, M Sheth, E Shinbrot, L Wang, M Wang, DA Wheeler, L Xi, F Zhao, J Hess, EL Appelbaum, M Bailey, MG Cordes, L Ding, CC Fronick, LA Fulton, RS Fulton, C Kandoth, ER Mardis, MD McLellan, CA Miller, HK Schmidt, RK Wilson, D Crain, E Curley, J Gardner, K Lau, D Mallery, S Morris, J Paulauskis, R Penny, C Shelton, T Shelton, M Sherman, E Thompson, P Yena, J Bowen, JM Gastier-Foster, M Gerken, KM Leraas, TM Lichtenberg, NC Ramirez, L Wise, E Zmuda, N Corcoran, T Costello, C Hovens, AL Carvalho, AC De Carvalho, JH Fregnani, A Longatto-Filho, RM Reis, C Scapulatempo-Neto, HCS Silveira, DO Vidal, A Burnette, J Eschbacher, B Hermes, A Noss, R Singh, ML Anderson, PD Castro, M Ittmann, D Huntsman, B Kohl, X Le, R Thorp, C Andry, ER Duffy, V Lyadov, O Paklina, G Setdikova, A Shabunin, M Tavobilov, C McPherson, R Warnick, R Berkowitz, D Cramer, C Feltmate, N Horowitz, A Kibel, M Muto, CP Raut, A Malykh, JS Barnholtz-Sloan, W Barrett, K Devine, J Fulop, QT Ostrom, K Shimmel, Y Wolinsky, AE Sloan, A De Rose, F Giuliante, M Goodman, BY Karlan, CH Hagedorn, J Eckman, J Harr, J Myers, K Tucker, LA Zach, B Deyarmin, H Hu, L Kvecher, C Larson, RJ Mural, S Somiari, A Vicha, T Zelinka, J Bennett, M Iacocca, B Rabeno, P Swanson, M Latour, L Lacombe, B Têtu, A Bergeron, M McGraw, SM Staugaitis, J Chabot, H Hibshoosh, A Sepulveda, T Su, T Wang, O Potapova, O Voronina, L Desjardins, O Mariani, S Roman-Roman, X Sastre, MH Stern, F Cheng, S Signoretti, A Berchuck, D Bigner, E Lipp, J Marks, S McCall, R McLendon, A Secord, A Sharp, M Behera, DJ Brat, A Chen, K Delman, S Force, F Khuri, K Magliocca, S Maithel, JJ Olson, T Owonikoko, A Pickens, S Ramalingam, DM Shin, G Sica, EG Van Meir, H Zhang, W Eijckenboom, A Gillis, E Korpershoek, L Looijenga, W Oosterhuis, H Stoop, KE Van Kessel, EC Zwarthoff, C Calatozzolo, L Cuppini, S Cuzzubbo, F DiMeco, G Finocchiaro, L Mattei, A Perin, B Pollo, C Chen, J Houck, P Lohavanichbutr, A Hartmann, C Stoehr, R Stoehr, H Taubert, S Wach, B Wullich, W Kycler, D Murawa, M Wiznerowicz, K Chung, WJ Edenfield, J Martin, E Baudin, G Bubley, R Bueno, A De Rienzo, WG Richards, S Kalkanis, T Mikkelsen, H Noushmehr, L Scarpace, N Girard, M Aymerich, E Campo, E Giné, AL Guillermo, N Van Bang, PT Hanh, BD Phu, Y Tang, H Colman, K Evason, PR Dottino, JA Martignetti, H Gabra, H Juhl, T Akeredolu, S Stepa, D Hoon, K Ahn, KJ Kang, F Beuschlein, A Breggia, M Birrer, D Bell, M Borad, AH Bryce, E Castle, V Chandan, J Cheville, JA Copland, M Farnell, T Flotte, N Giama, T Ho, M Kendrick, JP Kocher, K Kopp, C Moser, D Nagorney, D O’Brien, BP O’Neill, T Patel, G Petersen, F Que, M Rivera, L Roberts, R Smallridge, T Smyrk, M Stanton, RH Thompson, M Torbenson, JD Yang, L Zhang, F Brimo, JA Ajani, AMA Gonzalez, C Behrens, J Bondaruk, R Broaddus, B Czerniak, B Esmaeli, J Fujimoto, J Gershenwald, C Guo, AJ Lazar, C Logothetis, F Meric-Bernstam, C Moran, L Ramondetta, D Rice, A Sood, P Tamboli, T Thompson, P Troncoso, A Tsao, I Wistuba, C Carter, L Haydu, P Hersey, V Jakrot, H Kakavand, R Kefford, K Lee, G Long, G Mann, M Quinn, R Saw, R Scolyer, K Shannon, A Spillane, J Stretch, M Synott, J Thompson, J Wilmott, H Al-Ahmadie, TA Chan, R Ghossein, A Gopalan, DA Levine, V Reuter, S Singer, B Singh, NV Tien, T Broudy, C Mirsaidi, P Nair, P Drwiega, J Miller, J Smith, H Zaren, JW Park, NP Hung, E Kebebew, WM Linehan, AR Metwalli, K Pacak, PA Pinto, M Schiffman, LS Schmidt, CD Vocke, N Wentzensen, R Worrell, H Yang, M Moncrieff, C Goparaju, J Melamed, H Pass, N Botnariuc, I Caraman, M Cernat, I Chemencedji, A Clipca, S Doruc, G Gorincioi, S Mura, M Pirtac, I Stancul, D Tcaciuc, M Albert, I Alexopoulou, A Arnaout, J Bartlett, J Engel, S Gilbert, J Parfitt, H Sekhon, G Thomas, DM Rassl, RC Rintoul, C Bifulco, R Tamakawa, W Urba, N Hayward, H Timmers, A Antenucci, F Facciolo, G Grazi, M Marino, R Merola, R De Krijger, AP Gimenez-Roqueplo, A Piché, S Chevalier, G McKercher, K Birsoy, G Barnett, C Brewer, C Farver, T Naska, NA Pennell, D Raymond, C Schilero, K Smolenski, F Williams, C Morrison, JA Borgia, MJ Liptay, M Pool, CW Seder, K Junker, L Omberg, M Dinkin, G Manikhas, D Alvaro, MC Bragazzi, V Cardinale, G Carpino, E Gaudio, D Chesla, S Cottingham, M Dubina, F Moiseenko, R Dhanasekaran, KF Becker, KP Janssen, J Slotta-Huspenina, MH Abdel-Rahman, D Aziz, S Bell, CM Cebulla, A Davis, R Duell, JB Elder, J Hilty, B Kumar, J Lang, NL Lehman, R Mandt, P Nguyen, R Pilarski, K Rai, L Schoenfield, K Senecal, P Wakely, P Hansen, R Lechan, J Powers, A Tischler, WE Grizzle, KC Sexton, A Kastl, J Henderson, S Porten, J Waldmann, M Fassnacht, SL Asa, D Schadendorf, M Couce, M Graefen, H Huland, G Sauter, T Schlomm, R Simon, P Tennstedt, O Olabode, M Nelson, O Bathe, PR Carroll, JM Chan, P Disaia, P Glenn, RK Kelley, CN Landen, J Phillips, M Prados, J Simko, K Smith-McCune, S VandenBerg, K Roggin, A Fehrenbach, A Kendler, S Sifri, R Steele, A Jimeno, F Carey, I Forgie, M Mannelli, M Carney, B Hernandez, B Campos, C Herold-Mende, C Jungk, A Unterberg, A Von Deimling, A Bossler, J Galbraith, L Jacobus, M Knudson, T Knutson, D Ma, M Milhem, R Sigmund, AK Godwin, R Madan, HG Rosenthal, C Adebamowo, SN Adebamowo, A Boussioutas, D Beer, T Giordano, AM Mes-Masson, F Saad, T Bocklage, L Landrum, R Mannel, K Moore, K Moxley, R Postier, J Walker, R Zuna, M Feldman, F Valdivieso, R Dhir, J Luketich, EMM Pinero, M Quintero-Aguilo, CG Carlotti, JS Dos Santos, R Kemp, A Sankarankuty, D Tirapelli, J Catto, K Agnew, E Swisher, J Creaney, B Robinson, CS Shelley, EM Godwin, S Kendall, C Shipman, C Bradford, T Carey, A Haddad, J Moyer, L Peterson, M Prince, L Rozek, G Wolf, R Bowman, KM Fong, I Yang, R Korst, WK Rathmell, JL Fantacone-Campbell, JA Hooke, AJ Kovatich, CD Shriver, J DiPersio, B Drake, R Govindan, S Heath, T Ley, B Van Tine, P Westervelt, MA Rubin, JI Lee, ND Aredes, A Mariamidze. “Comprehensive Molecular Characterization of the Hippo Signaling Pathway in Cancer”, Cell Reports 25(5):1304-1317.e5 (2018). doi:10.1016/j.celrep.2018.10.001 |
|
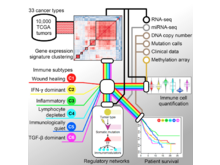
V Thorsson, DL Gibbs, SD Brown, D Wolf, DS Bortone, TH Ou Yang, E Porta-Pardo, GF Gao, CL Plaisier, JA Eddy, E Ziv, AC Culhane, EO Paull, IKA Sivakumar, AJ Gentles, R Malhotra, F Farshidfar, A Colaprico, JS Parker, LE Mose, NS Vo, J Liu, Y Liu, J Rader, V Dhankani, SM Reynolds, R Bowlby, A Califano, AD Cherniack, D Anastassiou, D Bedognetti, Y Mokrab, AM Newman, A Rao, K Chen, A Krasnitz, H Hu, TM Malta, H Noushmehr, CS Pedamallu, S Bullman, AI Ojesina, A Lamb, W Zhou, H Shen, TK Choueiri, JN Weinstein, J Guinney, J Saltz, RA Holt, CS Rabkin, The Cancer Genome Atlas Research Network (incl. N Gehlenborg), AJ Lazar, JS Serody, EG Demicco, ML Disis, BG Vincent, I Shmulevich. “The Immune Landscape of Cancer”, Immunity 48(4):812-830.e14 (2018). doi:10.1016/j.immuni.2018.03.023 |
|
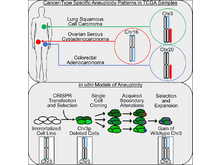
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “Genomic and Functional Approaches to Understanding Cancer Aneuploidy”, Cancer Cell 33(4):676-689.e3 (2018). doi:10.1016/j.ccell.2018.03.007 |
|
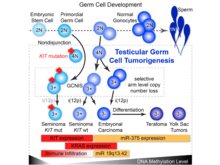
H Shen, J Shih, DP Hollern, L Wang, R Bowlby, SK Tickoo, V Thorsson, AJ Mungall, Y Newton, AM Hegde, J Armenia, F Sánchez-Vega, J Pluta, LC Pyle, R Mehra, VE Reuter, G Godoy, J Jones, CS Shelley, DR Feldman, DO Vidal, D Lessel, T Kulis, FM Cárcano, KM Leraas, TM Lichtenberg, D Brooks, AD Cherniack, J Cho, DI Heiman, K Kasaian, M Liu, MS Noble, L Xi, H Zhang, W Zhou, JC ZenKlusen, CM Hutter, I Felau, J Zhang, N Schultz, G Getz, M Meyerson, JM Stuart, The Cancer Genome Atlas Research Network (incl. N Gehlenborg), R Akbani, DA Wheeler, PW Laird, KL Nathanson, VK Cortessis, KA Hoadley. “Integrated Molecular Characterization of Testicular Germ Cell Tumors”, Cell Reports 23(11):3392-3406 (2018). doi:10.1016/j.celrep.2018.05.039 |

|
M Seiler, S Peng, AA Agrawal, J Palacino, T Teng, P Zhu, PG Smith, The Cancer Genome Atlas Research Network (incl. N Gehlenborg), S Buonamici, L Yu. “Somatic Mutational Landscape of Splicing Factor Genes and Their Functional Consequences across 33 Cancer Types”, Cell Reports 23(1):282-296.e4 (2018). doi:10.1016/j.celrep.2018.01.088 |
|
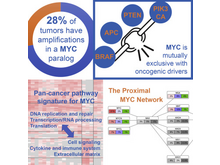
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “Pan-cancer Alterations of the MYC Oncogene and Its Proximal Network across the Cancer Genome Atlas”, Cell Systems 6(3):282-300.e2 (2018). doi:10.1016/j.cels.2018.03.003 |
|
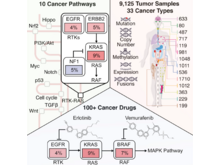
F Sanchez-Vega, M Mina, J Armenia, WK Chatila, A Luna, KC La, S Dimitriadoy, DL Liu, HS Kantheti, S Saghafinia, D Chakravarty, F Daian, Q Gao, MH Bailey, WW Liang, SM Foltz, I Shmulevich, L Ding, Z Heins, A Ochoa, B Gross, J Gao, H Zhang, R Kundra, C Kandoth, I Bahceci, L Dervishi, U Dogrusoz, W Zhou, H Shen, PW Laird, GP Way, CS Greene, H Liang, Y Xiao, C Wang, A Iavarone, AH Berger, TG Bivona, AJ Lazar, GD Hammer, T Giordano, LN Kwong, G McArthur, C Huang, AD Tward, MJ Frederick, F McCormick, M Meyerson, The Cancer Genome Atlas Research Network (incl. N Gehlenborg), EM Van Allen, AD Cherniack, G Ciriello, C Sander, N Schultz,. “Oncogenic Signaling Pathways in The Cancer Genome Atlas”, Cell 173(2):321-337.e10 (2018). doi:10.1016/j.cell.2018.03.035 |
|
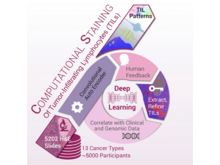
J Saltz, R Gupta, L Hou, T Kurc, P Singh, V Nguyen, D Samaras, KR Shroyer, T Zhao, R Batiste, J Van Arnam, The Cancer Genome Atlas Research Network (incl. N Gehlenborg), I Shmulevich, AUK Rao, AJ Lazar, A Sharma, V Thorsson. “Spatial Organization and Molecular Correlation of Tumor-Infiltrating Lymphocytes Using Deep Learning on Pathology Images”, Cell Reports 23(1):181-193.e7 (2018). doi:10.1016/j.celrep.2018.03.086 |
|
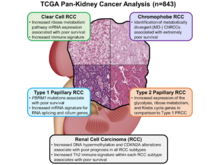
CJ Ricketts, AA De Cubas, H Fan, CC Smith, M Lang, E Reznik, R Bowlby, EA Gibb, R Akbani, R Beroukhim, DP Bottaro, TK Choueiri, RA Gibbs, AK Godwin, S Haake, AA Hakimi, EP Henske, JJ Hsieh, TH Ho, RS Kanchi, B Krishnan, DJ Kwiatkowski, W Liu, MJ Merino, GB Mills, J Myers, ML Nickerson, VE Reuter, LS Schmidt, CS Shelley, H Shen, B Shuch, S Signoretti, R Srinivasan, P Tamboli, G Thomas, BG Vincent, CD Vocke, DA Wheeler, L Yang, WY Kim, AG Robertson, The Cancer Genome Atlas Research Network (incl. N Gehlenborg), PT Spellman, WK Rathmell, WM Linehan. “The Cancer Genome Atlas Comprehensive Molecular Characterization of Renal Cell Carcinoma”, Cell Reports 23(1):313-326.e5 (2018). doi:10.1016/j.celrep.2018.03.075 |
|
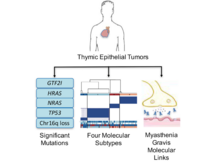
M Radovich, CR Pickering, I Felau, G Ha, H Zhang, H Jo, KA Hoadley, P Anur, J Zhang, M McLellan, R Bowlby, T Matthew, L Danilova, AM Hegde, J Kim, MDM Leiserson, G Sethi, C Lu, M Ryan, X Su, AD Cherniack, G Robertson, R Akbani, P Spellman, JN Weinstein, DN Hayes, B Raphael, T Lichtenberg, K Leraas, JC Zenklusen, The Cancer Genome Atlas Network (incl. N Gehlenborg), J Fujimoto, C Scapulatempo-Neto, AL Moreira, D Hwang, J Huang, M Marino, R Korst, G Giaccone, Y Gokmen-Polar, S Badve, A Rajan, P Ströbel, N Girard, MS Tsao, A Marx, AS Tsao, PJ Loehrer. “The Integrated Genomic Landscape of Thymic Epithelial Tumors”, Cancer Cell 33(2):244-258.e10 (2018). doi:10.1016/j.ccell.2018.01.003 |

|
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “Molecular Characterization and Clinical Relevance of Metabolic Expression Subtypes in Human Cancers”, Cell Reports 23(1):255-269.e4 (2018). doi:10.1016/j.celrep.2018.03.077 |

|
C Nobre, N Gehlenborg, H Coon, A Lex. “Lineage: Visualizing Multivariate Clinical Data in Genealogy Graphs”, IEEE Transactions on Visualization and Computer Graphics 25(3):1543-1558 (2018). doi:10.1109/TVCG.2018.2811488 |
|
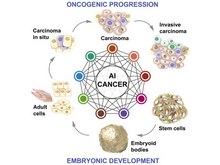
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation”, Cell 173(2):338-354.e15 (2018). doi:10.1016/j.cell.2018.03.034 |
|
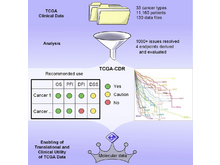
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics”, Cell 173(2):400-416.e11 (2018). doi:10.1016/j.cell.2018.02.052 |

|
F Lekschas, B Bach, P Kerpedjiev, N Gehlenborg, H Pfister. “HiPiler: Visual Exploration Of Large Genome Interaction Matrices With Interactive Small Multiples”, IEEE Transactions on Visualization and Computer Graphics 24(1):522-531 (2018). doi:10.1109/TVCG.2017.2745978 |

|
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “A Pan-Cancer Analysis Reveals High-Frequency Genetic Alterations in Mediators of Signaling by the TGF-β Superfamily”, Cell Systems 7(4):422-437.e7 (2018). doi:10.1016/j.cels.2018.08.010 |
|
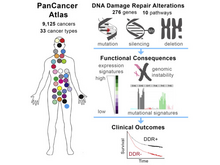
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas”, Cell Reports 23(1):239-254.e6 (2018). doi:10.1016/j.celrep.2018.03.076 |

|
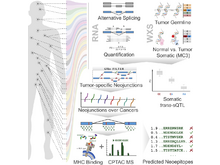
P Kerpedjiev, N Abdennur, F Lekschas, C McCallum, K Dinkla, H Strobelt, JM Luber, SB Ouellette, A Azhir, N Kumar, J Hwang, S Lee, BH Alver, H Pfister, LA Mirny, PJ Park, N Gehlenborg. “HiGlass: web-based visual exploration and analysis of genome interaction maps”, Genome Biology 19(1):125 (2018). doi:10.1186/s13059-018-1486-1 |
|
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “Comprehensive Analysis of Alternative Splicing Across Tumors from 8,705 Patients”, Cancer Cell 34(2):211-224.e6 (2018). doi:10.1016/j.ccell.2018.07.001 |
|
RG Jayasinghe, S Cao, Q Gao, MC Wendl, NS Vo, SM Reynolds, Y Zhao, H Climente-González, S Chai, F Wang, R Varghese, M Huang, WW Liang, MA Wyczalkowski, S Sengupta, Z Li, SH Payne, D Fenyö, JH Miner, MJ Walter, The Cancer Genome Atlas Research Network (incl. N Gehlenborg), B Vincent, E Eyras, K Chen, I Shmulevich, F Chen, L Ding. “Systematic Analysis of Splice-Site-Creating Mutations in Cancer”, Cell Reports 23(1):270-281.e3 (2018). doi:10.1016/j.celrep.2018.03.052 |
|
K Huang, RJ Mashl, Y Wu, DI Ritter, J Wang, C Oh, M Paczkowska, S Reynolds, MA Wyczalkowski, N Oak, AD Scott, M Krassowski, AD Cherniack, KE Houlahan, R Jayasinghe, LB Wang, DC Zhou, D Liu, S Cao, YW Kim, A Koire, JF McMichael, V Hucthagowder, TB Kim, A Hahn, C Wang, MD McLellan, F Al-Mulla, KJ Johnson, The Cancer Genome Atlas Research Network (incl. N Gehlenborg), O Lichtarge, PC Boutros, B Raphael, AJ Lazar, W Zhang, MC Wendl, R Govindan, S Jain, D Wheeler, S Kulkarni, JF Dipersio, J Reimand, F Meric-Bernstam, K Chen, I Shmulevich, SE Plon, F Chen, L Ding. “Pathogenic Germline Variants in 10,389 Adult Cancers”, Cell 173(2):355-370.e14 (2018). doi:10.1016/j.cell.2018.03.039 |

|
KA Hoadley, C Yau, T Hinoue, DM Wolf, AJ Lazar, E Drill, R Shen, AM Taylor, AD Cherniack, V Thorsson, R Akbani, R Bowlby, CK Wong, M Wiznerowicz, F Sanchez-Vega, AG Robertson, BG Schneider, MS Lawrence, H Noushmehr, TM Malta, JM Stuart, CC Benz, PW Laird, SJ Caesar-Johnson, JA Demchok, I Felau, M Kasapi, ML Ferguson, CM Hutter, HJ Sofia, R Tarnuzzer, Z Wang, L Yang, JC Zenklusen, JJ Zhang, S Chudamani, J Liu, L Lolla, R Naresh, T Pihl, Q Sun, Y Wan, Y Wu, J Cho, T DeFreitas, S Frazer, N Gehlenborg, G Getz, DI Heiman, J Kim, MS Lawrence, P Lin, S Meier, MS Noble, G Saksena, D Voet, H Zhang, B Bernard, N Chambwe, V Dhankani, T Knijnenburg, R Kramer, K Leinonen, Y Liu, M Miller, S Reynolds, I Shmulevich, V Thorsson, W Zhang, R Akbani, BM Broom, AM Hegde, Z Ju, RS Kanchi, A Korkut, J Li, H Liang, S Ling, W Liu, Y Lu, GB Mills, KS Ng, A Rao, M Ryan, J Wang, JN Weinstein, J Zhang, A Abeshouse, J Armenia, D Chakravarty, WK Chatila, I De Bruijn, J Gao, BE Gross, ZJ Heins, R Kundra, K La, M Ladanyi, A Luna, MG Nissan, A Ochoa, SM Phillips, E Reznik, F Sanchez-Vega, C Sander, N Schultz, R Sheridan, SO Sumer, Y Sun, BS Taylor, J Wang, H Zhang, P Anur, M Peto, P Spellman, C Benz, JM Stuart, CK Wong, C Yau, DN Hayes, JS Parker, MD Wilkerson, A Ally, M Balasundaram, R Bowlby, D Brooks, R Carlsen, E Chuah, N Dhalla, R Holt, SJM Jones, K Kasaian, D Lee, Y Ma, MA Marra, M Mayo, RA Moore, AJ Mungall, K Mungall, AG Robertson, S Sadeghi, JE Schein, P Sipahimalani, A Tam, N Thiessen, K Tse, T Wong, AC Berger, R Beroukhim, AD Cherniack, C Cibulskis, SB Gabriel, GF Gao, G Ha, M Meyerson, SE Schumacher, J Shih, MH Kucherlapati, RS Kucherlapati, S Baylin, L Cope, L Danilova, MS Bootwalla, PH Lai, DT Maglinte, DJ Van Den Berg, DJ Weisenberger, JT Auman, S Balu, T Bodenheimer, C Fan, KA Hoadley, AP Hoyle, SR Jefferys, CD Jones, S Meng, PA Mieczkowski, LE Mose, AH Perou, CM Perou, J Roach, Y Shi, JV Simons, T Skelly, MG Soloway, D Tan, U Veluvolu, H Fan, T Hinoue, PW Laird, H Shen, W Zhou, M Bellair, K Chang, K Covington, CJ Creighton, H Dinh, HV Doddapaneni, LA Donehower, J Drummond, RA Gibbs, R Glenn, W Hale, Y Han, J Hu, V Korchina, S Lee, L Lewis, W Li, X Liu, M Morgan, D Morton, D Muzny, J Santibanez, M Sheth, E Shinbrot, L Wang, M Wang, DA Wheeler, L Xi, F Zhao, J Hess, EL Appelbaum, M Bailey, MG Cordes, L Ding, CC Fronick, LA Fulton, RS Fulton, C Kandoth, ER Mardis, MD McLellan, CA Miller, HK Schmidt, RK Wilson, D Crain, E Curley, J Gardner, K Lau, D Mallery, S Morris, J Paulauskis, R Penny, C Shelton, T Shelton, M Sherman, E Thompson, P Yena, J Bowen, JM Gastier-Foster, M Gerken, KM Leraas, TM Lichtenberg, NC Ramirez, L Wise, E Zmuda, N Corcoran, T Costello, C Hovens, AL Carvalho, AC De Carvalho, JH Fregnani, A Longatto-Filho, RM Reis, C Scapulatempo-Neto, HCS Silveira, DO Vidal, A Burnette, J Eschbacher, B Hermes, A Noss, R Singh, ML Anderson, PD Castro, M Ittmann, D Huntsman, B Kohl, X Le, R Thorp, C Andry, ER Duffy, V Lyadov, O Paklina, G Setdikova, A Shabunin, M Tavobilov, C McPherson, R Warnick, R Berkowitz, D Cramer, C Feltmate, N Horowitz, A Kibel, M Muto, CP Raut, A Malykh, JS Barnholtz-Sloan, W Barrett, K Devine, J Fulop, QT Ostrom, K Shimmel, Y Wolinsky, AE Sloan, A De Rose, F Giuliante, M Goodman, BY Karlan, CH Hagedorn, J Eckman, J Harr, J Myers, K Tucker, LA Zach, B Deyarmin, H Hu, L Kvecher, C Larson, RJ Mural, S Somiari, A Vicha, T Zelinka, J Bennett, M Iacocca, B Rabeno, P Swanson, M Latour, L Lacombe, B Têtu, A Bergeron, M McGraw, SM Staugaitis, J Chabot, H Hibshoosh, A Sepulveda, T Su, T Wang, O Potapova, O Voronina, L Desjardins, O Mariani, S Roman-Roman, X Sastre, MH Stern, F Cheng, S Signoretti, A Berchuck, D Bigner, E Lipp, J Marks, S McCall, R McLendon, A Secord, A Sharp, M Behera, DJ Brat, A Chen, K Delman, S Force, F Khuri, K Magliocca, S Maithel, JJ Olson, T Owonikoko, A Pickens, S Ramalingam, DM Shin, G Sica, EG Van Meir, H Zhang, W Eijckenboom, A Gillis, E Korpershoek, L Looijenga, W Oosterhuis, H Stoop, KE Van Kessel, EC Zwarthoff, C Calatozzolo, L Cuppini, S Cuzzubbo, F DiMeco, G Finocchiaro, L Mattei, A Perin, B Pollo, C Chen, J Houck, P Lohavanichbutr, A Hartmann, C Stoehr, R Stoehr, H Taubert, S Wach, B Wullich, W Kycler, D Murawa, M Wiznerowicz, K Chung, WJ Edenfield, J Martin, E Baudin, G Bubley, R Bueno, A De Rienzo, WG Richards, S Kalkanis, T Mikkelsen, H Noushmehr, L Scarpace, N Girard, M Aymerich, E Campo, E Giné, AL Guillermo, N Van Bang, PT Hanh, BD Phu, Y Tang, H Colman, K Evason, PR Dottino, JA Martignetti, H Gabra, H Juhl, T Akeredolu, S Stepa, D Hoon, K Ahn, KJ Kang, F Beuschlein, A Breggia, M Birrer, D Bell, M Borad, AH Bryce, E Castle, V Chandan, J Cheville, JA Copland, M Farnell, T Flotte, N Giama, T Ho, M Kendrick, JP Kocher, K Kopp, C Moser, D Nagorney, D O’Brien, BP O’Neill, T Patel, G Petersen, F Que, M Rivera, L Roberts, R Smallridge, T Smyrk, M Stanton, RH Thompson, M Torbenson, JD Yang, L Zhang, F Brimo, JA Ajani, AMA Gonzalez, C Behrens, O Bondaruk, R Broaddus, B Czerniak, B Esmaeli, J Fujimoto, J Gershenwald, C Guo, AJ Lazar, C Logothetis, F Meric-Bernstam, C Moran, L Ramondetta, D Rice, A Sood, P Tamboli, T Thompson, P Troncoso, A Tsao, I Wistuba, C Carter, L Haydu, P Hersey, V Jakrot, H Kakavand, R Kefford, K Lee, G Long, G Mann, M Quinn, R Saw, R Scolyer, K Shannon, A Spillane, J Stretch, M Synott, J Thompson, J Wilmott, H Al-Ahmadie, TA Chan, R Ghossein, A Gopalan, DA Levine, V Reuter, S Singer, B Singh, NV Tien, T Broudy, C Mirsaidi, P Nair, P Drwiega, J Miller, J Smith, H Zaren, JW Park, NP Hung, E Kebebew, WM Linehan, AR Metwalli, K Pacak, PA Pinto, M Schiffman, LS Schmidt, CD Vocke, N Wentzensen, R Worrell, H Yang, M Moncrieff, C Goparaju, J Melamed, H Pass, N Botnariuc, I Caraman, M Cernat, I Chemencedji, A Clipca, S Doruc, G Gorincioi, S Mura, M Pirtac, I Stancul, D Tcaciuc, M Albert, I Alexopoulou, A Arnaout, J Bartlett, J Engel, S Gilbert, J Parfitt, H Sekhon, G Thomas, DM Rassl, RC Rintoul, C Bifulco, R Tamakawa, W Urba, N Hayward, H Timmers, A Antenucci, F Facciolo, G Grazi, M Marino, R Merola, R De Krijger, AP Gimenez-Roqueplo, A Piché, S Chevalier, G McKercher, K Birsoy, G Barnett, C Brewer, C Farver, T Naska, NA Pennell, D Raymond, C Schilero, K Smolenski, F Williams, C Morrison, JA Borgia, MJ Liptay, M Pool, CW Seder, K Junker, L Omberg, M Dinkin, G Manikhas, D Alvaro, MC Bragazzi, V Cardinale, G Carpino, E Gaudio, D Chesla, S Cottingham, M Dubina, F Moiseenko, R Dhanasekaran, KF Becker, KP Janssen, J Slotta-Huspenina, MH Abdel-Rahman, D Aziz, S Bell, CM Cebulla, A Davis, R Duell, JB Elder, J Hilty, B Kumar, J Lang, NL Lehman, R Mandt, P Nguyen, R Pilarski, K Rai, L Schoenfield, K Senecal, P Wakely, P Hansen, R Lechan, J Powers, A Tischler, WE Grizzle, KC Sexton, A Kastl, J Henderson, S Porten, J Waldmann, M Fassnacht, SL Asa, D Schadendorf, M Couce, M Graefen, H Huland, G Sauter, T Schlomm, R Simon, P Tennstedt, O Olabode, M Nelson, O Bathe, PR Carroll, JM Chan, P Disaia, P Glenn, RK Kelley, CN Landen, J Phillips, M Prados, J Simko, K Smith-McCune, S VandenBerg, K Roggin, A Fehrenbach, A Kendler, S Sifri, R Steele, A Jimeno, F Carey, I Forgie, M Mannelli, M Carney, B Hernandez, B Campos, C Herold-Mende, C Jungk, A Unterberg, A Von Deimling, A Bossler, J Galbraith, L Jacobus, M Knudson, T Knutson, D Ma, M Milhem, R Sigmund, AK Godwin, R Madan, HG Rosenthal, C Adebamowo, SN Adebamowo, A Boussioutas, D Beer, T Giordano, AM Mes-Masson, F Saad, T Bocklage, L Landrum, R Mannel, K Moore, K Moxley, R Postier, J Walker, R Zuna, M Feldman, F Valdivieso, R Dhir, J Luketich, EMM Pinero, M Quintero-Aguilo, CG Carlotti, JS Dos Santos, R Kemp, A Sankarankuty, D Tirapelli, J Catto, K Agnew, E Swisher, J Creaney, B Robinson, CS Shelley, EM Godwin, S Kendall, C Shipman, C Bradford, T Carey, A Haddad, J Moyer, L Peterson, M Prince, L Rozek, G Wolf, R Bowman, KM Fong, I Yang, R Korst, WK Rathmell, JL Fantacone-Campbell, JA Hooke, AJ Kovatich, CD Shriver, J DiPersio, B Drake, R Govindan, S Heath, T Ley, B Van Tine, P Westervelt, MA Rubin, JI Lee, ND Aredes, A Mariamidze. “Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer”, Cell 173(2):291-304.e6 (2018). doi:10.1016/j.cell.2018.03.022 |
|
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “Integrative Molecular Characterization of Malignant Pleural Mesothelioma”, Cancer Discovery 8(12):1548-1565 (2018). doi:10.1158/2159-8290.CD-18-0804 |
|
Z Ge, JS Leighton, Y Wang, X Peng, Z Chen, H Chen, Y Sun, F Yao, J Li, H Zhang, J Liu, CD Shriver, H Hu, The Cancer Genome Atlas Research Network (incl. N Gehlenborg), H Piwnica-Worms, L Ma, H Liang. “Integrated Genomic Analysis of the Ubiquitin Pathway across Cancer Types”, Cell Reports 23(1):213-226.e3 (2018). doi:10.1016/j.celrep.2018.03.047 |

|
Q Gao, WW Liang, SM Foltz, G Mutharasu, RG Jayasinghe, S Cao, WW Liao, SM Reynolds, MA Wyczalkowski, L Yao, L Yu, SQ Sun, The Fusion Analysis Working Group, The Cancer Genome Atlas Research Network (incl. N Gehlenborg), K Chen, AJ Lazar, RC Fields, MC Wendl, BA Van Tine, R Vij, F Chen, M Nykter, I Shmulevich, L Ding, . “Driver Fusions and Their Implications in the Development and Treatment of Human Cancers”, Cell Reports 23(1):227-238.e3 (2018). doi:10.1016/j.celrep.2018.03.050 |
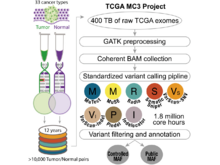
|
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines”, Cell Systems 6(3):271-281.e7 (2018). doi:10.1016/j.cels.2018.03.002 |
|
HS Chiu, S Somvanshi, E Patel, TW Chen, VP Singh, B Zorman, SL Patil, Y Pan, SS Chatterjee, The Cancer Genome Atlas Research Network (incl. N Gehlenborg), AK Sood, PH Gunaratne, P Sumazin. “Pan-Cancer Analysis of lncRNA Regulation Supports Their Targeting of Cancer Genes in Each Tumor Context”, Cell Reports 23(1):297-312.e12 (2018). doi:10.1016/j.celrep.2018.03.064 |
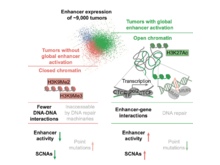
|
H Chen, C Li, X Peng, Z Zhou, JN Weinstein, The Cancer Genome Atlas Research Network (incl. N Gehlenborg), H Liang. “A Pan-Cancer Analysis of Enhancer Expression in Nearly 9000 Patient Samples”, Cell 173(2):386-399.e12 (2018). doi:10.1016/j.cell.2018.03.027 |
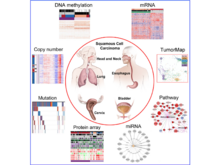
|
JD Campbell, C Yau, R Bowlby, Y Liu, K Brennan, H Fan, AM Taylor, C Wang, V Walter, R Akbani, LA Byers, CJ Creighton, C Coarfa, J Shih, AD Cherniack, O Gevaert, M Prunello, H Shen, P Anur, J Chen, H Cheng, DN Hayes, S Bullman, CS Pedamallu, AI Ojesina, S Sadeghi, KL Mungall, AG Robertson, C Benz, A Schultz, RS Kanchi, CM Gay, A Hegde, L Diao, J Wang, W Ma, P Sumazin, HS Chiu, TW Chen, P Gunaratne, L Donehower, JS Rader, R Zuna, H Al-Ahmadie, AJ Lazar, ER Flores, KY Tsai, JH Zhou, AK Rustgi, E Drill, R Shen, CK Wong, The Cancer Genome Atlas Research Network (incl. N Gehlenborg), JM Stuart, PW Laird, KA Hoadley, JN Weinstein, M Peto, CR Pickering, Z Chen, C Van Waes. “Genomic, Pathway Network, and Immunologic Features Distinguishing Squamous Carcinomas”, Cell Reports 23(1):194-212.e6 (2018). doi:10.1016/j.celrep.2018.03.063 |

|
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “A Comprehensive Pan-Cancer Molecular Study of Gynecologic and Breast Cancers”, Cancer Cell 33(4):690-705.e9 (2018). doi:10.1016/j.ccell.2018.03.014 |

|
M Behrisch, R Krueger, F Lekschas, T Schreck, N Gehlenborg, H Pfister. “Visual Pattern-Driven Exploration of Big Data”, Proceedings of International Symposium on Big Data Visual and Immersive Analytics (BDVA) (2018). doi:10.1109/BDVA.2018.8534028 |

|
J Aerts, N Gehlenborg, GE Marai, KK Nieselt. “Visualization of Biological Data - Crossroads (Dagstuhl Seminar 18161)”, Schloss Dagstuhl – Leibniz-Zentrum für Informatik 32-71 (2018). |
2017 | |

|
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma”, Cancer Cell 32(2):185-203.e13 (2017). doi:10.1016/j.ccell.2017.07.007 |
|
F Lekschas and N Gehlenborg. “SATORI: A System for Ontology-Guided Visual Exploration of Biomedical Data Repositories”, Bioinformatics 34(7):1200-1207 (2017). doi:10.1093/bioinformatics/btx739 |
|
M Kern, A Lex, N Gehlenborg, CR Johnson. “Interactive Visual Exploration And Refinement Of Cluster Assignments”, BMC Bioinformatics 18(1):406 (2017). doi:10.1186/s12859-017-1813-7 |

|
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “Integrated genomic characterization of oesophageal carcinoma”, Nature 541(7636):169-175 (2017). doi:10.1038/nature20805 |

|
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “Integrated genomic and molecular characterization of cervical cancer”, Nature 543(7645):378-384 (2017). doi:10.1038/nature21386 |
|
AD Cherniack, H Shen, V Walter, C Stewart, BA Murray, R Bowlby, X Hu, S Ling, RA Soslow, RR Broaddus, RE Zuna, G Robertson, PW Laird, R Kucherlapati, GB Mills, The Cancer Genome Atlas Research Network (incl. N Gehlenborg), JN Weinstein, J Zhang, R Akbani, DA Levine. “Integrated Molecular Characterization of Uterine Carcinosarcoma”, Cancer Cell 31(3):411-423 (2017). doi:10.1016/j.ccell.2017.02.010 |

|
L Fishbein, I Leshchiner, V Walter, L Danilova, AG Robertson, AR Johnson, TM Lichtenberg, BA Murray, HK Ghayee, T Else, S Ling, SR Jefferys, AA De Cubas, B Wenz, E Korpershoek, AL Amelio, L Makowski, WK Rathmell, AP Gimenez-Roqueplo, TJ Giordano, SL Asa, AS Tischler, The Cancer Genome Atlast Research Network (incl. N Gehlenborg), K Pacak, KL Nathanson, MD Wilkerson. “Comprehensive Molecular Characterization of Pheochromocytoma and Paraganglioma”, Cancer Cell 31(2):181-193 (2017). doi:10.1016/j.ccell.2017.01.001 |

|
JR Conway, A Lex, N Gehlenborg. “UpSetR: An R Package for the Visualization of Intersecting Sets and their Properties”, Bioinformatics 33(18):2938-2940 (2017). doi:10.1093/bioinformatics/btx364 |
|
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “Comprehensive and Integrated Genomic Characterization of Adult Soft Tissue Sarcomas”, Cell 171(4):950-965.e28 (2017). doi:10.1016/j.cell.2017.10.014 |
2016 | |

|
R Hourieh, H Stitz, N Gehlenborg, M Streit. “TaCo: Comparative Visualization of Large Tabular Data”, EuroVis 2016 - Posters (2016). doi:10.2312/EURP.20161149 |
|
AK Manrai, CJ Patel, N Gehlenborg, NP Tatonetti, JPA Ionnaidis, IS Kohane. “Methods to enhance the reproducibility of precision medicine”, Biocomputing 2016: Proceedings of the Pacific Symposium (2016). doi:10.1142/9789814749411_0017 |
|
|
JD Campbell, A Alexandrov, J Kim, J Wala, AH Berger, CS Pedamallu, SA Shukla, G Guo, AN Brooks, BA Murray, M Imielinski, X Hu, S Ling, R Akbani, M Rosenberg, C Cibulskis, A Ramachandran, EA Collisson, DJ Kwiatkowski, MS Lawrence, JN Weinstein, RGW Verhaak, CJ Wu, PS Hammerman, AD Cherniack, G Getz, The Cancer Genome Atlas Research Network (incl. N Gehlenborg), MN Artyomov, R Schreiber, R Govindan, M Meyerson. “Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas”, Nature Genetics 48(6):607-616 (2016). doi:10.1038/ng.3564 |
|
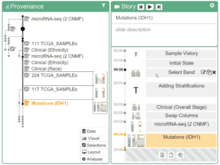
|
S Gratzl, A Lex, N Gehlenborg, N Cosgrove, M Streit. “From Visual Exploration to Storytelling and Back Again”, Computer Graphics Forum 35(3):491-500 (2016). doi:10.1111/cgf.12925 |
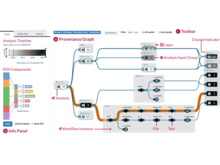
|
H Stitz, S Luger, M Streit, N Gehlenborg. “AVOCADO: Visualization of Workflow-Derived Data Provenance for Reproducible Biomedical Research”, Computer Graphics Forum 35(3):481-490 (2016). doi:10.1111/cgf.12924 |

|
M Ceccarelli, FP Barthel, TM Malta, TS Sabedot, SR Salama, BA Murray, O Morozova, Y Newton, A Radenbaugh, SM Pagnotta, S Anjum, J Wang, G Manyam, P Zoppoli, S Ling, AA Rao, M Grifford, AD Cherniack, H Zhang, L Poisson, CG Carlotti, DPC Tirapelli, A Rao, T Mikkelsen, CC Lau, WKA Yung, R Rabadan, J Huse, DJ Brat, NL Lehman, JS Barnholtz-Sloan, S Zheng, K Hess, G Rao, M Meyerson, R Beroukhim, L Cooper, R Akbani, M Wrensch, D Haussler, KD Aldape, PW Laird, DH Gutmann, The Cancer Genome Atlas Research Network (incl. N Gehlenborg), H Noushmehr, A Iavarone, RGW Verhaak. “Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma”, Cell 164(3):550-563 (2016). doi:10.1016/j.cell.2015.12.028 |
2015 | |

|
M Streit and N Gehlenborg. “Points of View: Temporal data”, Nature Methods 12(2):97-97 (2015). doi:10.1038/nmeth.3262 |
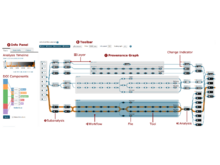
|
S Luger, H Stitz, S Gratzl, N Gehlenborg, M Streit. “Interactive Visualization of Provenance Graphs for Reproducible Biomedical Research”, Poster Compendium of the IEEE Conference on Information Visualization (InfoVis’ 15) (2015). |
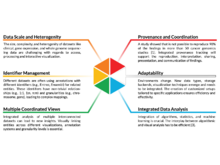
|
S Gratzl, N Gehlenborg, A Lex, H Strobelt, C Partl, M Streit. “Caleydo Web: An Integrated Visual Analysis Platform for Biomedical Data”, Poster Compendium of the IEEE Conference on Information Visualization (InfoVis’ 15) (2015). |

|
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “Comprehensive and Integrative Genomic Characterization of Diffuse Lower Grade Gliomas”, New England Journal of Medicine 372(26):2481-2498 (2015). doi:10.1056/NEJMoa1402121 |
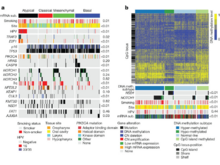
|
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “Comprehensive Genomic Characterization of Head and Neck Squamous Cell Carcinomas”, Nature 517(7536):576-582 (2015). doi:10.1038/nature14129 |

|
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “The Molecular Taxonomy of Primary Prostate Cancer”, Cell 163(4):1011-1025 (2015). doi:10.1016/j.cell.2015.10.025 |
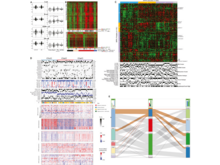
|
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “Genomic Classification of Cutaneous Melanoma”, Cell 161(7):1681-1696 (2015). doi:10.1016/j.cell.2015.05.044 |
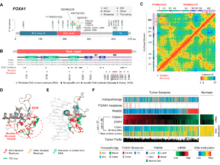
|
G Ciriello, ML Gatza, AH Beck, MD Wilkerson, SK Rhie, A Pastore, H Zhang, M McLellan, C Yau, C Kandoth, R Bowlby, H Shen, S Hayat, R Fieldhouse, SC Lester, GMK Tse, RE Factor, LC Collins, KH Allison, YY Chen, K Jensen, NB Johnson, S Oesterreich, GB Mills, AD Cherniack, G Robertson, C Benz, C Sander, PW Laird, KA Hoadley, TA King, The Cancer Genome Atlas Research Network (incl. N Gehlenborg), CM Perou. “Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer”, Cell 163(2):506-519 (2015). doi:10.1016/j.cell.2015.09.033 |
2014 | |
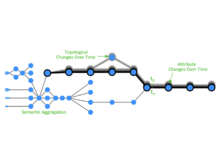
|
H Stitz, S Gratzl, S Luger, N Gehlenborg, M Streit. “Transparent layering for visualizing dynamic graphs using the flip book metaphor”, Poster Compendium of the IEEE VIS Conference. IEEE (2014). |
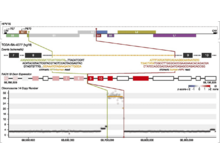
|
M Parfenov, CS Pedamallu, N Gehlenborg, SS Freeman, L Danilova, CA Bristow, S Lee, AG Hadjipanayis, EV Ivanova, MD Wilkerson, A Protopopov, L Yang, S Seth, X Song, J Tang, X Ren, J Zhang, A Pantazi, N Santoso, AW Xu, H Mahadeshwar, DA Wheeler, RI Haddad, J Jung, AI Ojesina, N Issaeva, WG Yarbrough, DN Hayes, JR Grandis, AK El-Naggar, M Meyerson, PJ Park, L Chin, JG Seidman, PS Hammerman, R Kucherlapati, The Cancer Genome Atlas Research Network. “Characterization of HPV and Host-Genome Interactions in Primary Head and Neck Cancers”, Proceedings of the National Academy of Sciences 111(43):15544-15549 (2014). doi:10.1073/pnas.1416074111 |
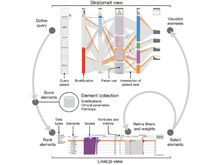
|
M Streit, A Lex, S Gratzl, C Partl, D Schmalstieg, H Pfister, PJ Park, N Gehlenborg. “Guided Visual Exploration of Genomic Stratifications in Cancer”, Nature Methods 11(9):884-885 (2014). doi:10.1038/nmeth.3088 |
|
A Lex and N Gehlenborg. “Points of View: Sets and Intersections”, Nature Methods 11(8):779 (2014). doi:10.1038/nmeth.3033 |
|
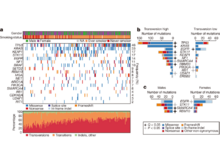
|
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “Comprehensive molecular profiling of lung adenocarcinoma”, Nature 511(7511):543-550 (2014). doi:10.1038/nature13385 |
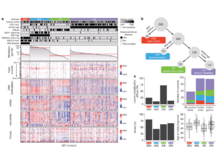
|
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “Comprehensive Molecular Characterization of Gastric Adenocarcinoma”, Nature 513(7517):202-209 (2014). doi:10.1038/nature13480 |
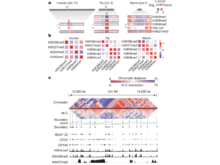
|
JWK Ho, YL Jung, T Liu, BH Alver, S Lee, K Ikegami, KA Sohn, A Minoda, MY Tolstorukov, A Appert, SCJ Parker, T Gu, A Kundaje, NC Riddle, E Bishop, TA Egelhofer, SS Hu, AA Alekseyenko, A Rechtsteiner, D Asker, JA Belsky, SK Bowman, QB Chen, RAJ Chen, DS Day, Y Dong, AC Dose, X Duan, CB Epstein, S Ercan, EA Feingold, F Ferrari, JM Garrigues, N Gehlenborg, PJ Good, P Haseley, D He, M Herrmann, MM Hoffman, TE Jeffers, PV Kharchenko, P Kolasinska-Zwierz, CV Kotwaliwale, N Kumar, SA Langley, EN Larschan, I Latorre, MW Libbrecht, X Lin, R Park, MJ Pazin, HN Pham, A Plachetka, B Qin, YB Schwartz, N Shoresh, P Stempor, A Vielle, C Wang, CM Whittle, H Xue, RE Kingston, JH Kim, BE Bernstein, AF Dernburg, V Pirrotta, MI Kuroda, WS Noble, TD Tullius, M Kellis, DM MacAlpine, S Strome, SCR Elgin, XS Liu, JD Lieb, J Ahringer, GH Karpen, PJ Park. “Comparative analysis of metazoan chromatin organization”, Nature 512(7515):449-452 (2014). doi:10.1038/nature13415 |
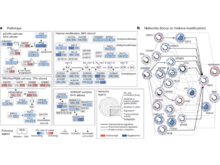
|
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “Comprehensive Molecular Characterization of Urothelial Bladder Carcinoma”, Nature 507(7492):315-322 (2014). doi:10.1038/nature12965 |
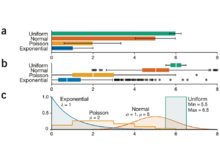
|
M Streit and N Gehlenborg. “Points of View: Bar Charts and Box Plots”, Nature Methods 11(2):117 (2014). doi:10.1038/nmeth.2807 |
|
A Lex, N Gehlenborg, H Strobelt, R Vuillemot, H Pfister. “UpSet: Visualization of Intersecting Sets”, IEEE Transactions on Visualization and Computer Graphics 20(12):1983-1992 (2014). doi:10.1109/TVCG.2014.2346248 |
|
|
S Carpendale, M Chen, D Evanko, N Gehlenborg, C Görg, L Hunter, F Rowland, MA Storey, H Strobelt. “Ontologies in Biological Data Visualization”, IEEE Computer Graphics and Applications 34(2):8-15 (2014). doi:10.1109/MCG.2014.33 |
|
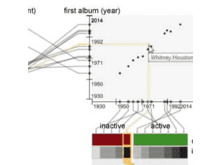
|
S Gratzl, N Gehlenborg, A Lex, H Pfister, M Streit. “Domino: Extracting, Comparing, and Manipulating Subsets across Multiple Tabular Datasets”, IEEE Transactions on Visualization and Computer Graphics 20(12):2023-2032 (2014). doi:10.1109/TVCG.2014.2346260 |
|
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “Integrated Genomic Characterization of Papillary Thyroid Carcinoma”, Cell 159(3):676-690 (2014). doi:10.1016/j.cell.2014.09.050
|
|

|
KA Hoadley, C Yau, DM Wolf, AD Cherniack, D Tamborero, S Ng, MDM Leiserson, B Niu, MD McLellan, V Uzunangelov, J Zhang, C Kandoth, R Akbani, H Shen, L Omberg, A Chu, AA Margolin, LJ van’t Veer, N Lopez-Bigas, PW Laird, BJ Raphael, L Ding, AG Robertson, LA Byers, GB Mills, JN Weinstein, C Van Waes, Z Chen, EA Collisson, The Cancer Genome Atlas Research Network (incl. N Gehlenborg), CC Benz, CM Perou, JM Stuart. “Multi-platform integration of 12 cancer types reveals cell-of-origin classes with distinct molecular signatures”, Cell 158(4):929-944 (2014). doi:10.1016/j.cell.2014.06.049 |
|
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “The Somatic Genomic Landscape of Chromophobe Renal Cell Carcinoma”, Cancer Cell 26(3):319-330 (2014). doi:10.1016/j.ccr.2014.07.014 |
2013 | |
|
J de Ridder, T Abeel, M Michaut, VP Satagopam, N Gehlenborg. “Don’t Wear Your New Shoes (Yet): Taking the Right Steps to Become a Successful Principal Investigator”, PLoS Computational Biology 9(1):e1002834 (2013). doi:10.1371/journal.pcbi.1002834 |
|
|
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “Comprehensive molecular characterization of clear cell renal cell carcinoma”, Nature 499(7456):43-49 (2013). doi:10.1038/nature12222 |
|
|
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “The Cancer Genome Atlas Pan-Cancer analysis project”, Nature Genetics 45(10):1113-1120 (2013). doi:10.1038/ng.2764 |
|
|
S Gratzl, A Lex, N Gehlenborg, H Pfister, M Streit. “LineUp: Visual Analysis of Multi-Attribute Rankings”, IEEE Transactions on Visualization and Computer Graphics 19(12):2277-2286 (2013). doi:10.1109/TVCG.2013.173
|
|
|
L Yang, LJ Luquette, N Gehlenborg, R Xi, PS Haseley, CH Hsieh, C Zhang, X Ren, A Protopopov, L Chin, R Kucherlapati, C Lee, PJ Park, The Cancer Genome Research Atlas (incl. N Gehlenborg). “Diverse mechanisms of somatic structural variations in human cancer genomes”, Cell 153(4):919-929 (2013). doi:10.1016/j.cell.2013.04.010 |
|
|
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “The somatic genomic landscape of glioblastoma”, Cell 155(2):462-477 (2013). doi:10.1016/j.cell.2013.09.034 |
|
|
N Gehlenborg, MS Noble, G Getz, L Chin, PJ Park. “Nozzle: a report generation toolkit for data analysis pipelines”, Bioinformatics 29(8):1089-1091 (2013). doi:10.1093/bioinformatics/btt085 |
|
|
S Ho Sui, E Merrill, N Gehlenborg, P Haseley, I Sytchev, R Park, P Rocca-Serra, S Corlosquet, A Gonzalez-Beltran, E Maguire, O Hofmann, PJ Park, S Das, SA Sansone, W Hide. “The Stem Cell Commons: an exemplar for data integration in the biomedical domain driven by the ISA framework”, AMIA Joint Summits on Translational Science 2013:70 (2013). |
|

|
J de Ridder, T Abeel, M Michaut, VP Satagopam, N Gehlenborg. “Don’t Wear Your New Shoes (Yet): Taking the Right Steps to Become a Successful Principal Investigator”, PLoS Computational Biology 9(1):e1002834 (2013). doi:10.1371/journal.pcbi.1002834 |
2012 | |
|
N Gehlenborg and B Wong. “Points of View: Into the Third Dimension”, Nature Methods 9(9):851 (2012). doi:10.1038/nmeth.2151 |
|
|
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “Comprehensive genomic characterization of squamous cell lung cancers”, Nature 489(7417):519-525 (2012). doi:10.1038/nature11404 |
|
|
N Gehlenborg and B Wong. “Points of View: Power of the Plane”, Nature Methods 9(10):935 (2012). doi:10.1038/nmeth.2186 |
|
|
N Gehlenborg and B Wong. “Points of View: Networks”, Nature Methods 9(2):115 (2012). doi:10.1038/nmeth.1862 |
|
|
N Gehlenborg and B Wong. “Points of View: Integrating Data”, Nature Methods 9(4):315 (2012). doi:10.1038/nmeth.1944 |
|
|
N Gehlenborg and B Wong. “Points of View: Heat maps”, Nature Methods 9(3):213 (2012). doi:10.1038/nmeth.1902 |
|
|
N Gehlenborg and B Wong. “Points of View: Mapping Quantitative Data to Color”, Nature Methods 9(8):769 (2012). doi:10.1038/nmeth.2134 |
|
|
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “Comprehensive molecular characterization of human colon and rectal cancer”, Nature 499(7456):43-49 (2012). doi:10.1038/nature12222 |
|
|
The Cancer Genome Atlas Research Network (incl. N Gehlenborg). “Comprehensive molecular portraits of human breast tumours”, Nature 490(7418):61-70 (2012). doi:10.1038/nature11412 |
|

|
A Lex, M Streit, HJ Schulz, C Partl, D Schmalstieg, PJ Park, N Gehlenborg. “StratomeX: Visual Analysis of Large-Scale Heterogeneous Genomics Data for Cancer Subtype Characterization”, Computer Graphics Forum 31:1175-1184 (2012). doi:10.1111/j.1467-8659.2012.03110.x
|
|
J Caldas, N Gehlenborg, E Kettunen, A Faisal, M Rönty, AG Nicholson, S Knuutila, A Brazma, S Kaski. “ Data-Driven Information Retrieval in Heterogeneous Collections of Transcriptomics Data Links SIM2s to Malignant Pleural Mesothelioma”, Bioinformatics 28(2):246-253 (2012). doi:10.1093/bioinformatics/btr634 |
|
2011 | |
|
R Park, N Gehlenborg, PJ Park. “Seqeyes: A multi-scale interactive visualization tool for structural variations”, IEEE Symposium on Biological Data Visualization 2011 (2011). |
2010 | |
|
N Gehlenborg. “Visualization and Exploration of Transcriptomics Data”, PhD Thesis, University of Cambridge, United Kingdom (2010). |
|
|
N Gehlenborg, SI O’Donoghue, NS Baliga, A Goesmann, MA Hibbs, H Kitano, O Kohlbacher, H Neuweger, R Schneider, D Tenenbaum, AC Gavin. “Visualization of Omics Data for Systems Biology”, Nature Methods 7(S3):S56-S68 (2010). doi:10.1038/nmeth.1436 |
|
|
SI O’Donoghue, AC Gavin, N Gehlenborg, DS Goodsell, JK Hériché, CB Nielsen, C North, AJ Olson, JB Procter, DW Shattuck, T Walter, B Wong. “Visualizing biological data – now and in the future”, Nature Methods 7(S3):S2-S4 (2010). doi:10.1038/nmeth.f.301 |
|
|
J Peltonen, H Aidos, N Gehlenborg, A Brazma, S Kaski. “An Information Retrieval Perspective on Visualization of Gene Expression Data with Ontological Annotation”, 2010 IEEE International Conference on Acoustics, Speech and Signal Processing (2010). doi:10.1109/ICASSP.2010.5495665 |
|
2009 | |
|
D Hwang, IY Lee, H Yoo, N Gehlenborg, JH Cho, B Petritis, D Baxter, R Pitstick, R Young, D Spicer, ND Price, JG Hohmann, SJ DeArmond, GA Carlson, LE Hood. “A Systems Approach to Prion Disease”, Molecular Systems Biology 5(1):252 (2009). doi:10.1038/msb.2009.10 |
|
|
N Gehlenborg, D Hwang, IY Lee, H Yoo, D Baxter, B Petritis, R Pitstick, B Marzolf, SJ DeArmond, GA Carlson, L Hood. “The Prion Disease Database: A Comprehensive Transcriptome Resource for Systems Biology Research in Prion Diseases”, Database 2009 (2009). doi:10.1093/database/bap011 |
|
|
N Gehlenborg and A Brazma. “Visualization of Large Microarray Experiments with Space Maps”, BMC Bioinformatics 10(S13):O7 (2009). doi:10.1186/1471-2105-10-S13-O7 |
|
|
J Caldas, N Gehlenborg, A Faisal, A Brazma, S Kaski. “Probabilistic Retrieval and Visualization of Biologically Relevant Microarray Experiments”, Bioinformatics 25(12):i145-i153 (2009). doi:10.1093/bioinformatics/btp215 |
|
|
N Gehlenborg, W Yan, IY Lee, H Yoo, K Nieselt, D Hwang, R Aebersold, L Hood. “Prequips – An Extensible Software Platform for Integration, Visualization and Analysis of LC-MS/MS Proteomics Data”, Bioinformatics 25(5):682-683 (2009). doi:10.1093/bioinformatics/btp005 |
|
|
J Caldas, N Gehlenborg, A Faisal, A Brazma, S Kaski. “Probabilistic retrieval and visualization of biologically relevant microarray experiments”, Bioinformatics 25(12):i145-i153 (2009). doi:10.1093/bioinformatics/btp215 |
|
2008 | |
|
A Schmidt, N Gehlenborg, B Bodenmiller, LN Mueller, D Campbell, M Mueller, R Aebersold, B Domon. “An integrated, directed mass spectrometric approach for in-depth characterization of complex peptide mixtures”, Molecular & Cellular Proteomics 7(11):2138-2150 (2008). doi:10.1074/mcp.M700498-MCP200 |
|
|
M Corpas, N Gehlenborg, SC Janga, PE Bourne. “Ten Simple Rules for Organizing a Scientific Meeting”, PLoS Computational Biology 4(6):e1000080 (2008). doi:10.1371/journal.pcbi.1000080 |
|
|
L Peixoto, N Gehlenborg, SC Janga. “Highlights from the Fourth International Society for Computational Biology (ISCB) Student Council Symposium at the Sixteenth Annual International Conference on Intelligent Systems for Molecular Biology (ISMB)”, BMC Bioinformatics 9(S10):I1 (2008). doi:10.1186/1471-2105-9-S10-I1 |
|
2007 | |
|
N Gehlenborg, M Corpas, SC Janga. “Highlights from the Third International Society for Computational Biology (ISCB) Student Council Symposium at the Fifteenth Annual International Conference on Intelligent Systems for Molecular Biology (ISMB)”, BMC Bioinformatics 8(S8):I1 (2007). doi:10.1186/1471-2105-8-S8-I1 |
|
2006 | |
|
DH Hwang, N Gehlenborg, N Zhang, W Yan, E Yi, HK Lee, IY Lee, R Aebersold, L Hood. “A computational framework for systems biology approaches using LC-MS based comparative proteomics”, The Korea Genome Conference (2006). |
|
|
N Gehlenborg. “Visualization and Integration of Liquid Chromatography/Mass Spectrometry Proteomics Data”, Diplomarbeit (eq. Masters Thesis), University of Tübingen, Germany (2006). |
|
|
J Dietzsch, N Gehlenborg, K Nieselt. “Mayday – A Microarray Data Analysis Workbench”, Bioinformatics 22(8):1010-1012 (2006). doi:10.1093/bioinformatics/btl070 |
|
2005 | |
|
N Gehlenborg. “Visualisierung für DNA-Chip-Analysen”, in D. Spath, K. Haasis and D. Klumpp (Eds.), Aktuelle Trends in der Softwareforschung (Vol. 3), 259-270, IRB Fraunhofer Verlag, Stuttgart, Germany (2005). |
|
|
N Gehlenborg, J Dietzsch, K Nieselt. “A Framework for Visualization of Microarray Data and Integrated Meta Information”, Information Visualization 4(3):164-175 (2005). doi:10.1057/palgrave.ivs.9500094 |
|
2003 | |
|
N Gehlenborg. “CLIMP-Cluster-based Imputation of Missing Values in Microarray Data”, (2003). |